Figures & data
Figure 1. Promoter and terminator elements description and entropy index for the transcribed region of the 5S rDNA. A) The promoter of the 5S rDNA contains the elements BoxA, the Internal Element, and the BoxC, altogether called Internal Control Region (ICR). The RNA polymerase recognizes a poly dT as stop signal. B) The entropy index for the 5S rRNA using all the 91 sequences retrieved from the GenBank. However, due the fact that the first 43 nucleotides in Potamotrygon motoro, Potamotrygon falkneri and Paratrygon aiereba transcript region sequences were missing, these were assumed to be equal to those in Urotrygon.
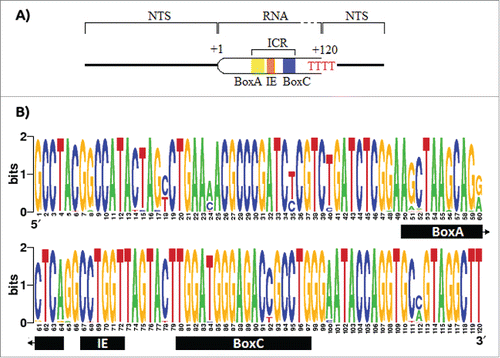
Table 1. List of the 5S rDNA type II gene sequences obtained from 14 elasmobranchs and 3 Petromyzontiformes species retrieve from the NCBI database.
Table 2. Contingency table of Chi-square to test the equality of the stop signal in all the elasmobranchii individuals studied here.
Figure 2. Evolutionary interpretation of the synapomorphic mutation events. (*)The synapomorphic mutation of the nucleotide 112 is absent in Galeocerdo cuvier. This might be due to the underrepresentation of this genus or a back mutation in that position.
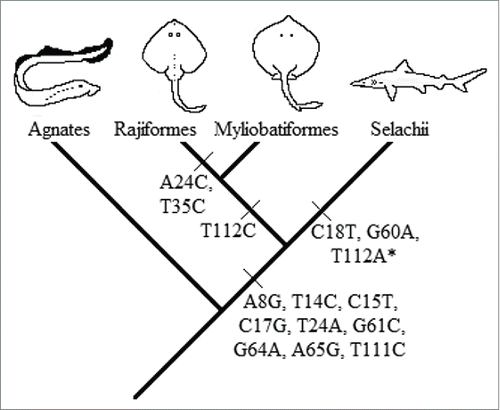
Figure 3. Secondary structure consensus of the 5S rRNA type II predicted by RNAalifold. The structure A) is proposed by Barciszewska et al.Citation48 which corresponds to the general model of 5S rRNA for eukaryotes. The structure B) is the consensus structure predicted for the 5S rRNA type II by RNAalifold. The main differences are those marked with red arrows: a) positions 7 and 112 with no pair in all sharks but G. cuvier; b) all the sequences fit well with the basepair 34C:41G and 33U:42A which reduce the loop C size; c) the bulge at the position 84 (with base dU in this study) proposed by Barciszewska et al.Citation48 is now located at the position 83 (dA). According to these authors the bulge located in the helix IV must be a purine; d) positions 80 and 96 are not joined in all the sequences.
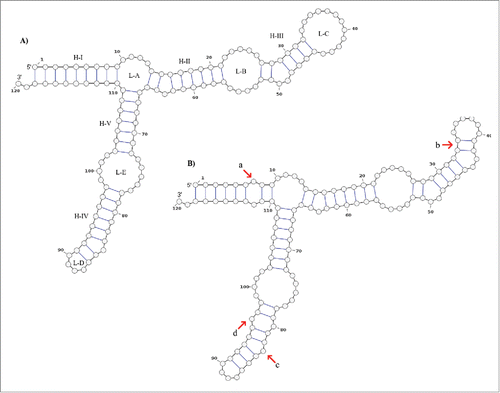
Figure 4. Secondary structural groups of the 5S rRNA type II defined by neighbor joining. The results show 3 different structural groups: α, β and γ. The group α is composed of structures that are similar to our consensus secondary structure of the 5S rRNA type II, while the groups β and γ are composed of structures different to our consensus. Red numbers represent the bootstrap values.
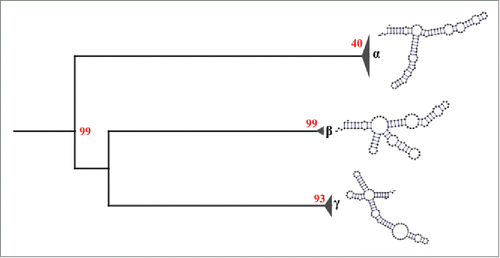
Table 3. Statistical results for the distribution of the microsallite region inside the NTS. Significant p-values for R. montagui and R. miraletus indicate that its distribution is regular. For other species the distribution is random.
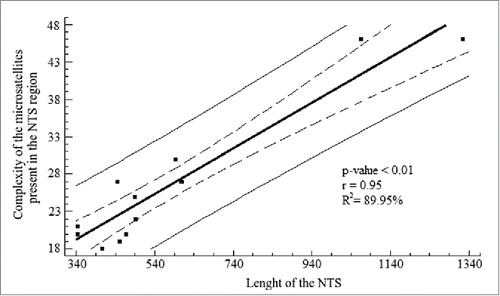
Figure 5. Linear regression analysis for the length of the NTS and the complexity of the microsatellites present in the NTS region. The analysis shows a high coefficient determination, which was significant. Solid line represents the linear regression according to the equation Y = 10.78 + 0.03(X) + e . Dashed lines represent the 95% confidence interval for the median value of Yi in any Xi . Thin lines are the prediction limits of 95% for new observations.