Figures & data
Figure 1. High levels of miRNA in pollen from different species. Plant MIR156a levels in fresh pollen collected by honey bee foragers (n = 32). Copy number is expressed per mg (A) and per 10pg total RNA (B).
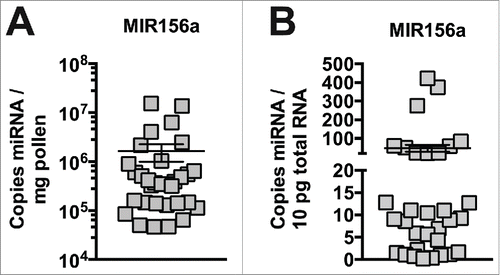
Table 1. Species Identification and MIR156a Levels of Honey Bee Collected Pollen. List of miRNA MIR156a levels in fresh pollen collected by honey bee foragers and the genus (and highest ranked species) of the plant of origin. Copy number is expressed per mg and per 10 pg total RNA.
Figure 2. Nurse and forager bees exhibit minimal levels of plant miRNA in abdominal tissue. Schematic of pollen consumption during honey bee life stages (based onCitation55,57) (A). Transcript levels of Vitellogenin relative to β-actin in midgut and abdominal tissue from nurse and forager bees taken directly from the hive (B). Plant MIR156a levels in midgut (C) or abdominal tissue (D) from nurse (n =8 ) or forager (n = 8) honey bees taken directly from the hive. Levels of endogenous mir277 in midgut (E) or abdominal tissue (F) from nurse and forager bees above. Plant MIR156a levels in midgut or abdominal tissue from newly eclosed honey bees from frames taken directly from the hive (G). Levels of endogenous mir277 in midgut or abdominal tissue from newly eclosed bees above (H). Copy number is expressed per 10pg total RNA. Data is expressed as Mean ± SEM, * signifies p < 0.05; ** signifies p < 0.01, n.s. signifies not significant, p > 0.05.
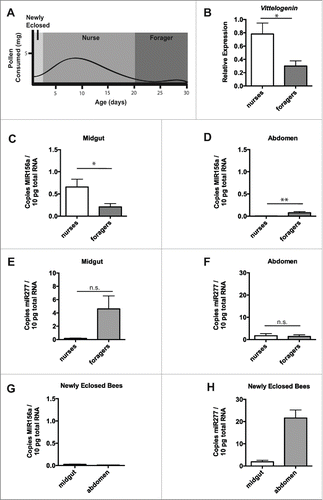
Figure 3. Negligible transfer of diet-derived miRNA to internal tissue after pollen ingestion. Plant miRNA MIR156a levels, expressed per 10pg total RNA, in honey bee midgut (A) and abdomen (B) after 24 hours of ingestion of sugar solution alone (n = 7) or sugar solution containing polyfloral pollen (n = 7) (after 48 hours of pollen-free diet). Endogenous honey bee miRNA mir277 levels in the honey bees above, expressed per 10pg total RNA, in honey bee midgut (C) and abdomen (D) after 24 hours of ingestion of sugar solution alone or sugar solution containing polyfloral pollen (after 48 hours of pollen-free diet). Copy number is expressed per 10pg total RNA. Data is expressed as Mean ± SEM, * signifies p < 0.05; ** signifies p < 0.01, n.s. signifies not significant, p > 0.05.
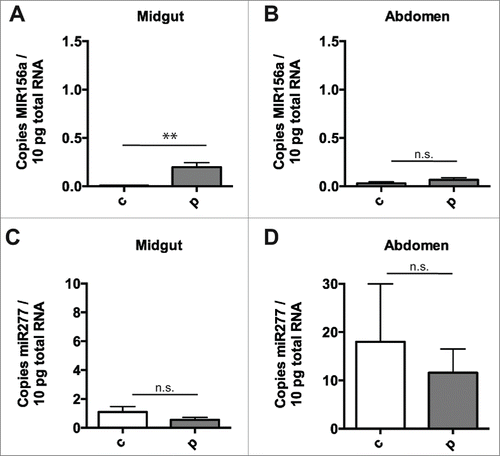
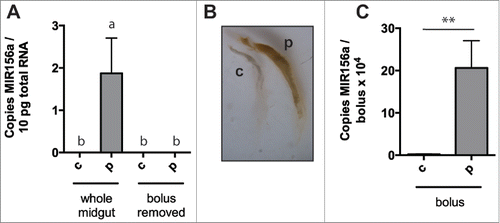
Figure 4. Negligible uptake of diet-derived miRNA by digestive tract epithelia after pollen ingestion. Plant miRNA MIR156a levels, expressed per 10pg total RNA, in whole honey bee midguts or midguts with food bolus removed after 24 hours of ingestion of sugar solution alone (n = 3) or sugar solution containing polyfloral pollen (n = 3) (after 48 hours of pollen-free diet) (A). Brightfield microscopic image of food boluses from midguts above (B). Plant miRNA MIR156a levels, expressed per bolus, in food boli removed from midguts above (n = 3) (C). Data is expressed as Mean ± SEM. In (A) a ≠ b, p < 0.05, the differences between b are not significant and in (B) * signifies p < 0.05; ** signifies p < 0.01.