Figures & data
Figure 1. Cmg1 specifically interacts with the RNA helicase Prp43. (A) Proteins were retrieved on IgG sepharose from extracts with or without tagged Cmg1, separated by SDS PAGE and stained with Coomassie. Inputs are shown on the left. Prp43 was identified in the Cmg1 eluate by mass spectrometry and Western blotting (bottom panel). The asterisk marks a background band also present in the control. (B) Recombinant Protein A-tagged Spp2 (as negative control) or Cmg1 were immobilised on IgG sepharose and incubated with purified GFP-tagged Prp43. After washing, co-purified GFP-Prp43 was eluted, then inputs and eluates were separated by SDS PAGE and analyzed by Coomassie staining. (C) Yeast two-hybrid analysis of Prp43 (fused to the GAL4 activation domain; GAL4-AD) was performed with full length (FL), the N-terminal G-patch domain (amino acids 1–85) or the C-terminus (amino acids 86–274) of Cmg1 (fused to the GAL4 binding domain; GAL4-BD), and Swm2 and Tgs1 as controls. The strains were spotted on plates not selecting (left) or selecting (right) for a yeast two-hybrid (Y2H) interaction. The domain structure of Cmg1, containing the G-patch domain and the domain of unidentified function (DUF4187) is shown at the top. (D) Protein A-tagged full length Prp43 or N- (92–767) or C- (1–657) terminally truncated versions of Prp43 were immobilised on IgG sepharose and incubated with MBP-tagged full length Cmg1 or Cmg1 1–85. After washing the beads, proteins were eluted, separated by SDS PAGE and analyzed by Commassie staining. Cmg1 and Cmg1 1–85 proteins co-precipitated with the different forms of Prp43 are indicated by the asterisk and double asterisk, respectively. For inputs of the binding experiments see Figure S1A.
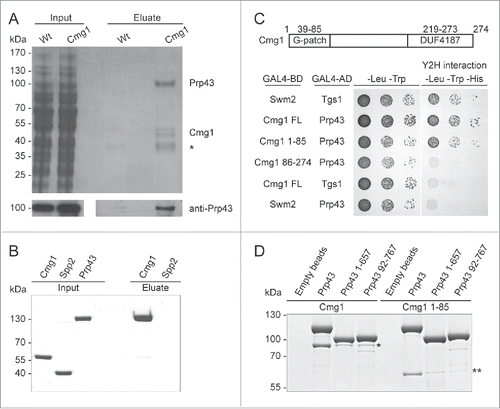
Figure 2. RNA binding and the ATPase activity of Prp43 are stimulated by Cmg1. (A) Fluorescence anisotropy titrations of fluorescein-labeled 11 nt RNA with Prp43 were performed with or without Cmg1 or the G-patch domain of Spp382 (Spp382 51–110). The Kd values are 60 ± 5 nM (Prp43 alone), 2.3 ± 1.6 nM (+Cmg1) and 2.5 ± 1.3 nM (+Spp382 51–110). (B) RNA-stimulated ATPase activity of Prp43 was measured in the absence or presence of Spp382 51–110 or Cmg1. The kcat values were 3.61 ± 0.14 s (Prp43 alone), 5.09 ± 0.26 s (+Cmg1) and 3.44 ± 0.13 s (+Spp382 51–110), the KM values 1.41 ± 0.14 μM (Prp43 alone), 0.23 ± 0.05 µM (+Cmg1) and 0.22 ± 0.04 µM (+Spp382 51–110).
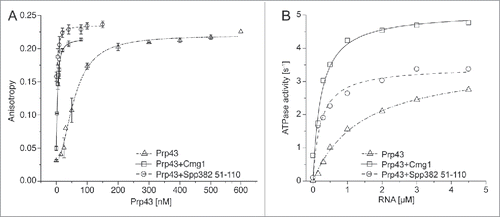
Figure 3. Cmg1 is localized in the cytoplasm and in the intermembrane space of mitochondria. (A) The localization of Cmg1-GFP (green in overlays) and Ssa2-RFP (panels on the left; red in overlay) or Mitotracker (panels on the right; red in overlay) and overlays are shown. Scale bar represents 5 µm for the main images. A 2-fold zoom of a representative cell is shown in each panel. (B) To analyze submitochondrial localization, yeast mitochondria were left untreated, swollen (Mitoplasts), or solubilised with 1% Triton X-100 (TX-100), then treated with Proteinase K (PK) where indicated, and subjected to SDS PAGE and Western blotting using antibodies against tagged Cmg1, the outer membrane protein Tom70, inner membrane proteins Mic10, Tim21 and Tim44. Note, Tim44 extrudes into the matrix, while a domain of Tim21 and the majority of Mic10 extends into the IMS. (C) Serial dilutions of Cmg1 overexpression (oex), overexpression of Cmg1 without its G-patch domain (86–274 oex), cmg1 deletion (Δcmg1) and wildtype strains were spotted for growth analysis with and without induction of apoptosis by treatment with acetic acid. Growth was documented after 2 days.
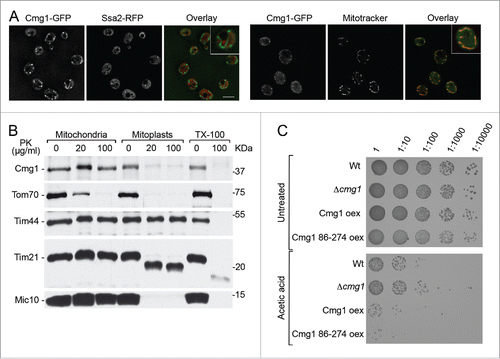
Figure 4. G-patch proteins compete for binding to Prp43. Recombinant Protein A-tagged G-patch domain of Spp382 was immobilised and incubated with GFP-Prp43 in the presence or absence of the indicated G-patch domains of Sqs1, Pxr1, Cmg1 or Spp2. Competitors were used in ratios of 1:1 and 5:1 compared to the immobilised G-patch domain of Spp382. Eluted GFP-Prp43 was quantified by fluorescence measurements. Data from three independent experiments are presented as mean +/− SEM.
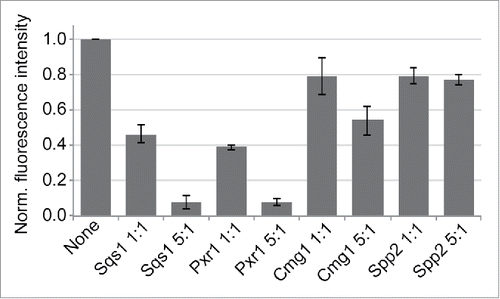
Figure 5. Overexpression of G-patch proteins leads to cellular relocalisation of Prp43. The localization of Prp43-GFP was analyzed by fluorescence microscopy in yeast cells overexpressing individual G-patch proteins or in control cells (Wt). The localization of the Prp43-GFP is shown on the left, the nucleolar marker Nop1-RFP in the middle, and the overlay with DAPI staining (blue) and a brightfield image is presented on the right. In the left and middle panels, cells are outlined with a white, dotted line. Nuclear foci containing GFP-Prp43 after overexpression of Spp382 are marked with arrowheads. The scale bar represents 5 µm.
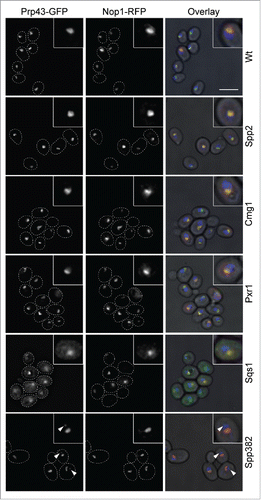
Figure 6. G-patch protein overexpression leads to defects in ribosome biogenesis. (A) Scheme of the 35S pre-rRNA transcript, which contains the sequences of the mature 18S, 5.8S and 25S rRNAs. (B) Total RNA was isolated from yeast cells with or without overexpression of the individual G-patch proteins indicated, RNA was separated by denaturing agarose gel electrophoresis and analyzed by Northern blotting using probes for the detection of rRNA precursors (indicated on the left). The 35S pre-rRNA is marked by an arrowhead. (C) Levels of the 35S rRNA precursor transcript in three independent experiments were quantified, normalized to the scR1 RNA (loading control) and the wildtype, and are presented as mean +/− SEM.
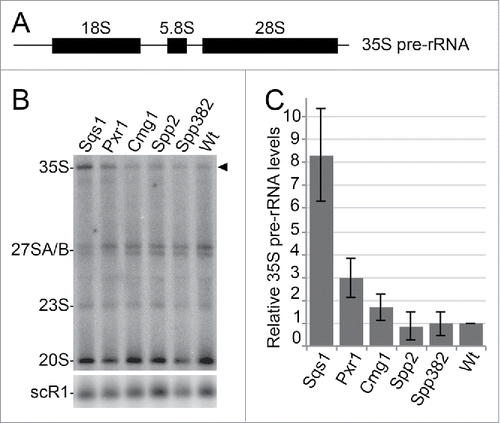
Figure 7. Scheme of the localisations of Prp43 and its G-patch protein cofactors. Prp43 is largely localized in nucleoli (No) where it is involved in ribosome synthesis and likely interacts with Pxr1/Gno1 and Sqs1/Pfa1. Prp43 is also found in nucleoplasm, where it acts in splicing with Spp382/Ntr1, and it is present in the cytoplasm, possibly binding Sqs1 or Cmg1. Dotted arrows indicate the exchange of Prp43 between its different cofactors.