Figures & data
Figure 1. Consensus model of the phylogeny of DNA 5mC / RNA m5C methyltransferases. Dnmt1 and Dnmt2/Dnmt3 enzymes have independent origin in the prokaryotic DNA methyltransferases. The putative precursor of the Dnmt2 family changed its substrate preference from DNA to RNA (indicated with *). Green shading denotes DNA methyltransferases, pink denotes RNA methyltransferases.
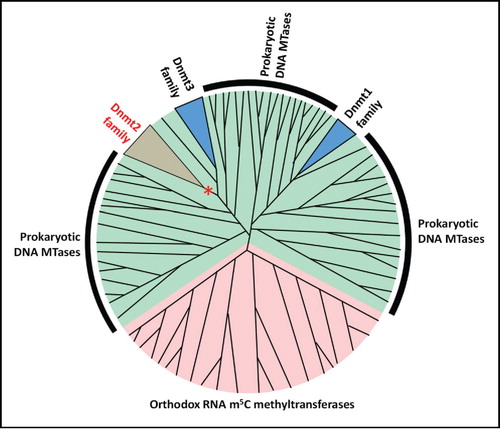
Figure 2. Structure of Entamoeba histolytica methyltransferase EhMeth (3QV2) and structure of tRNAAsp from S. cerevisiae (1VTQ) drawn in same scale. Dnmt2 is shown in schematic drawing in orange with SAM in yellow.
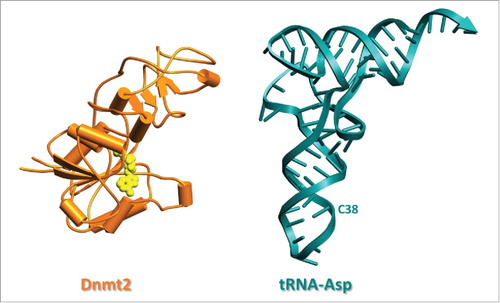
Figure 3. Substrate spectrum of Dnmt2. Dnmt2 has been shown to methylate tRNA-Asp and depending on the species tRNAGly, tRNAVal, and tRNAGlu. Whether other tRNAs, RNAs or DNA are methylated by this enzyme is an open question.
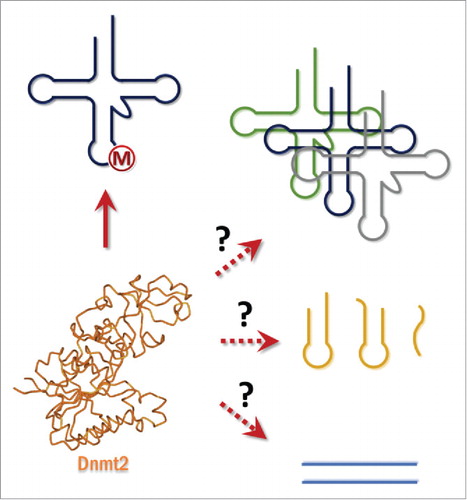
Table 1. Confirmed tRNA substrates of Dnmt2 enzymes.
Figure 4. Different views of DNMT2 in surface representation. The residues subjected to mutagenesis are colored according to their residual activity. The cofactor product S-adenosyl-l-homocysteine is colored blue. Panel A shows that residues which strongly interfere with catalysis (colored red here) cluster on the “front” face of the enzyme and surround a cleft that also contains the SAM binding pocket and the catalytic residues. Panel B represents a view of the “back side” of the enzyme after a 180° rotation of the view shown in panel A about the vertical axis. Reproduced fromCitation48 with permission. Copyright (2012) American Chemical Society.
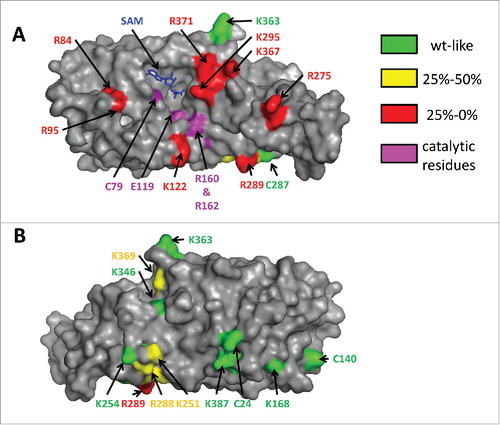
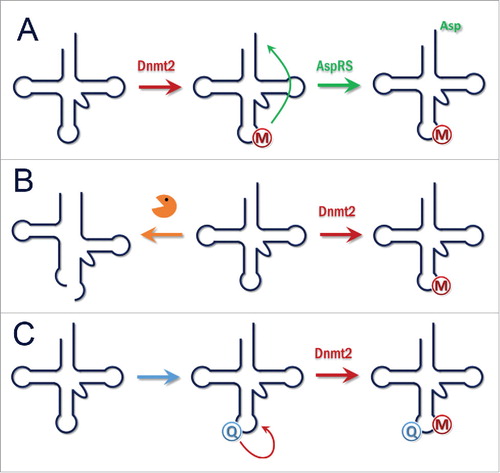
Figure 5. (A) Function of Dnmt2. Methylation of tRNAAspGUC enhances its charging by AspRS. This stimulates the translation of Asp-rich proteins. (B) Function of Dnmt2. Methylation of tRNAs protects them from cleavage into tRNA fragments and increases overall translation. Massive production of tRNA fragments after loss or downregulation of Dnmt2 affects Dicer dependent processing of small RNAs. C) Function of Dnmt2. Depending on nutrition or other signals, Queuosine is incorporated into tRNAAspGUC at position 34. This stimulates Dnmt2 mediated methylation and triggers downstream effects.