Figures & data
Figure 1. The illustration of Ago2-based RNA immunoprecipitation to immunoprecipated Ago2–RNA complexes followed by RNA sequencing (Ago2 RIP-seq) platform. (A) Western blot analysis shows the presence of Ago2 in Ago2 immunoprecipitation (IP) but not mouse IgG. (B) A scheme outlines Ago2 RIP-seq approach. A rabbit polyclonal anti-Ago2 and a mouse monoclonal anti-Ago2 antibody were used for the immunoprecipitation and Western blot analysis of Ago2, respectively. The normalization was performed by blotting the same samples with an antibody against β-actin. (C) The flow diagram outlines the various steps involved in RNA seq data analysis.
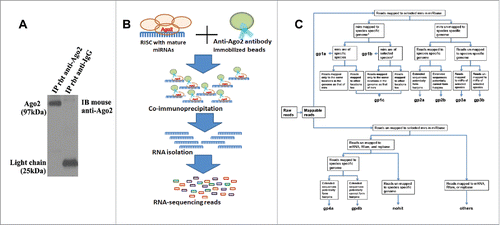
Figure 2. Characterization of Ago2-associated miRNomes in NPCs. Pie charts show the profiles of top 10 enriched Ago2-bound miRNAs in non-ischemic (A) and ischemic (B) NPCs. The bar-graph shows top 10 upregulated (C) and downregulated (D) miRNAs in ischemic NPCs. (E) Heat map diagram shows differential profiles of miRNAs in non-ischemic and ischemic NPCs. The color scale in (E)illustrates the relative expression level of miRNAs across all samples: red and green colors represent an expression level higher and lower, respectively, than the mean. Black represents median expression. (F) Bioinformatic analysis with Metacore and Targetscan softwares shows the signaling pathways related to highly enriched Ago2-bound miRNAs in ischemic NPCs. (G) QRT-PCR analysis validated top 3 upregulated enriched miRNAs and their target genes in NPCs after stroke. N = 3/group, **p < 0.01, ***p < 0.001. (H) Comparison of miRNA expression data between the Ago2-RIP and the miRNA microarray by means of Spearman correlation coefficient.
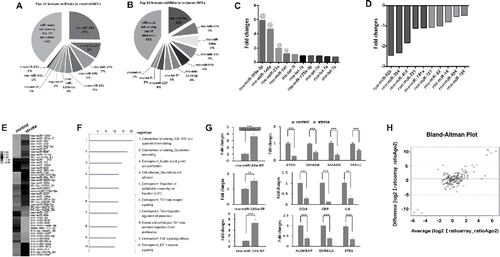
Figure 3. Analysis of isomiRs in non-ischemic and ischemic NPCs. (A) A bar-graph shows the distribution of modified mature miRNAs in the NPCs after stroke. (B) The percentage of normalized reads in the total library of top 20 most highly expressed isomiRs in the normal and ischemic NPCs. (C) Comparison of the shifted (bold red) and canonical (bold green) seeds of isomiR-142–5p_L+2R-1 and isomiR-187-5p_L-2R+3. Detailed sequence information and average frequency are provided in Supplementary Table S4. (D) Stem-loop TaqMan PCR and qRT-PCR analysis shows the upregulation of isomiR-142-5p_L+2R-1 and downregulation of isomiR-187-5p_L-2R+3 in ischemic NPCs. N = 3/group, **p < 0.01. (E and F) Venn diagrams show the overlapped genes putatively targeted by isomiR-142-5p_L+2R-1 or isomiR-187-5p_L-2R+3 (red) as well as by their parent canonical miR-142-5p and miR-187-5p (blue). Predictions were performed using the miRDB (left box) and TargetScan (right box) softwares.
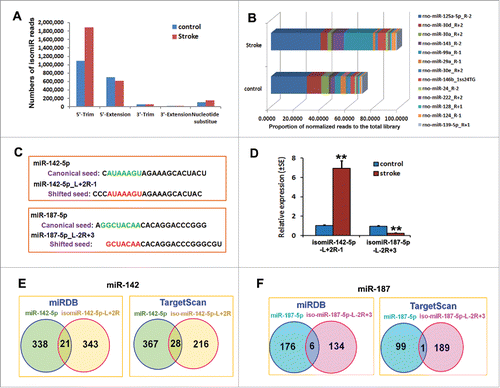
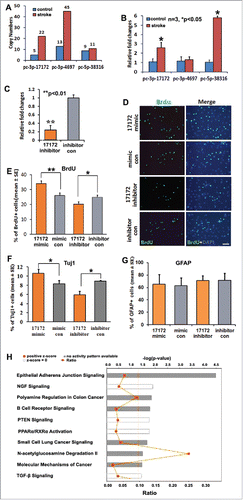
Figure 4. Novel miRNA pc-3p-17172 regulates neural progenitor cell proliferation and differentiation. (A) Ago2-RNA immunoprecipitation (RIP) shows the copy numbers of novel miRNAs in non-ischemic (control) and ischemic (MCAo) NPCs. (B) Quantitative RT-PCR data show the altered expression of novel miRNAs in non-ischemic (control) and ischemic (MCAo) NPCs. (C) The endogenous pc-3p-17172 expression was significantly knocked down in ischemic NPCs transfected by miScript inhibitor (pc-3p-17172 inhibitor) compared with NPCs transfected by inhibitor control (inhibitor con). Panel (D)shows the representative immunocytochemistry images of BrdU positive cells in ischemic NPCs transfected by pc-3p-17172 mimic, inhibitor or their corresponding controls. Panels E, (F)and (G)demonstrate quantitative data of BrdU (E), Tuj1 (F), GFAP (G) positive cells, respectively, in NPCs after transfection of pc-3p-17172 mimic or inhibitor. Panel (H)shows the most significantly enriched canonical signaling pathways associated with genes putatively targeted by pc-3p-17172. Data were analyzed by means of the IPA software. N = 3 for each group. * p < 0.05; ** p < 0.01 vs. the control group. Scale bar = 20µm.