Figures & data
Figure 1. Growth retardation in Hepatocytes specific Adar depleted mice. (A) Representative WB analysis of ADAR1 p110 and p150 isoforms in total cell lysates from the livers of 2- and 4-week old Adar Hep-KO mice and their littermate controls. Tubulin levels were used as loading controls. (B) Representative images of 4-week old Adar Hep-KO mice and livers compared to littermate controls. (C) Mean body weight of 4-week old Adar Hep-KO males (n = 17) and females (n = 10) compared to WT littermate controls. (D-E) Mean body weight and Liver to body mass ratio, repectively, of 2-week (n = 4) and 4-week old (n = 7) Adar Hep-KO mice compare to littermate controls.
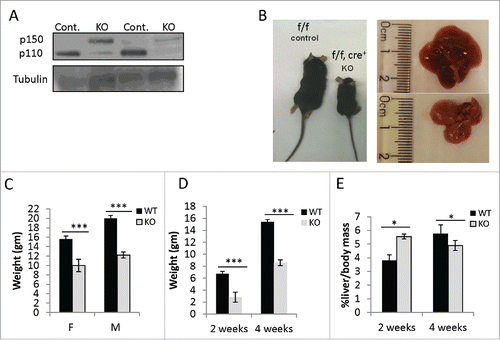
Figure 2. Adar Hep-KO mice show extensive liver damage with fibrogenesis. (A) Impaired level of liver enzymes detected in serum of 4-week old Adar Hep-KO mice: decreased levels of albumin, and increased levels of alkaline phosphatase (ALP), alanine aminotransferase (ALT), aspartate aminotransferase (AST) and lactate dehydrogenase (LDH) were detected in the Adar Hep-KO mice indicative of hepatocellular damage (n = 4). (B) H&E staining of representative liver sections from 2 and 4-week old Adar Hep-KO (KO) and WT control (WT) mice (n = 5). While the WT section exhibit normal characteristics the livers from the Adar Hep-KO mice show massive damages to the tissue (x40 magnification). (C) Analysis of representative liver section from 4-week old Adar Hep-KO and WT mice using specific staining to detect apoptosis (TUNEL assay, x10 magnification), compensatory proliferation (Ki67, x20 magnification), changes in liver architecture (Reticulin, x40 magnification) and hepatic fibrosis (Serius Red, x40 magnification).
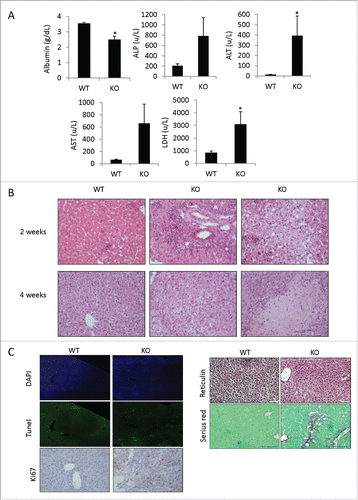
Figure 3. Adar Hep-KO livers show increased CD8+ cytotoxic T cells and granulocyte infiltration and a type-I interferon signature. (A-C) Flow cytometry analysis of non-parenchymal hepatic cells from 4-week old Adar Hep-KO and heterozygous littermate controls. After purifications of the cells erythrocytes were depleted and cells were immunostained for CD45R/B220, CD3, CD4, CD8, CD11b, Ly6G, and F4/80: (A) Analysis for the percentage of B cells (B220+CD3− and CD19+ gating), and for CD3+B220- T cells. (B) Depicting the CD3+ gate and further analysis for the percentages of CD4+ helper T cells and CD8+ cytotoxic T cells. (C) Analysis for the percentage of Ly6G+CD11b+ granulocytes (left panel) and F4/80+Ly6G- hepatic macrophages (right panel). Shown is a representative experiment out of n = 3 performed. (D) GSEA for biological functions and disease categories of the differentially expressed genes by Qiagen's IPA tool of 2 Adar Hep-KO mice versus 2 littermate controls (using 2FC cutoff). (E) Microarray mRNA expression levels of type-I interferons stimulated genes (ISGs) in 2 and 4-week old Adar Hep-KO vs. littermate control mice.
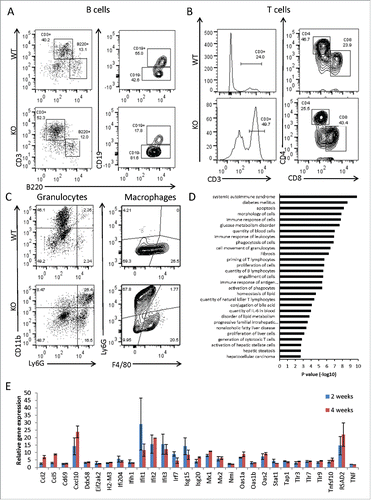
Figure 4. ADAR1 depleted HepG2 cells induce paracrine activation of primary hepatic stellate cells. HepG2 cells were either untreated (Unt) or transfected with siControl or siADAR1. (A) 48-hours post transfection the relative mRNA expression of ADAR1 p110 and p150 in HepG2 cells was measured by qPCR, normalized to HPRT. (B) WB analysis of total cell lysates using anti-ADAR1 and anti-actin (as loading control) specific Abs. (C) Primary hepatic stellate cells were incubated with conditioned medium collected from ADAR1 HepG2-KD and control cells, and 24 hours later the relative mRNA expression levels of αSMA and Col1A were measured using qPCR, normalized to HPRT. Bar graphs show mean ± SEM of n = 3 experiments.
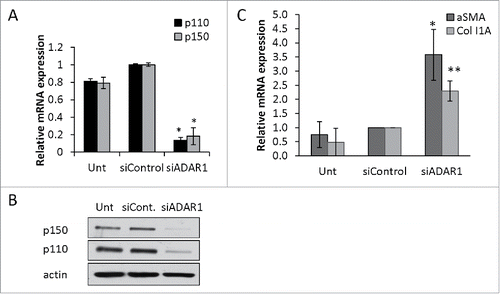
Figure 5. NFκB-dependent upregulation of IL6 and IL8 in ADAR1 HepG2-KD cells. (A) Relative mRNA expression of IL6 and IL8 in untreated, siControl and siADAR1 transfected HepG2 cells as determined by qPCR (normalized to HPRT). (B) Levels of IL6 in conditioned medium of siControl and siADAR1 transfected HepG2 cells as measured by ELISA kits. Results are representative of n = 3 experiments. (C) Untransfected, siControl and siADAR1 transfected HepG2 cells were co-transfected with pGL2-KB-Luciferase to assess the levels of NFkB activation. Luminescence values are shown relative to control Renila plasmid. (D) Relative expression of IL6 and IL8 mRNA in HepG2 cells following co-depletion of ADAR1 and p65. shP65 or shGFP control were co-transfected with siADAR1 or siControl. IL6 and IL8 expression levels were determined using qPCR. (E) The effect of IFNα treatment on mRNA expression levels of ADAR1 p110, ADAR1 p150, IL6 and IL8 in siADAR1 and siControl HepG2 transfected cells by qPCR, normalized to HPRT. Bar graphs show mean ± SEM of n = 3 experiments.
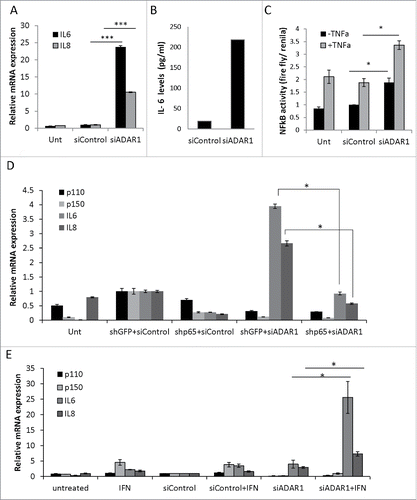
Figure 6. ADAR1 regulatory effect on NFκ(B)cascade is editing independent. (A) Relative IL6 mRNA expression levels normalized to HPRT as measured by qPCR, following co- transfection of HepG2 cells with siADAR1 and either one of the following 5 plasmids: p110 WT, p110 mutated in the editase domain (mut) p150WT, p150 mut and empty Flag plasmid. Transfection with siControl and empty Flag plasmid served as the negative control. Bar graphs show mean ± SEM of n = 3 experiments. (B) ADAR1-p110- GFP and ADAR1-p150-GFP plasmids were transfected into HepG2 cells. Co-IP of cell extracts using GFP-trap beads. The levels of IKKβ were detected using WB analysis with specific Abs. Expression of empty GFP vector served as negative control. (C) Co-IP of cell extracts using anti-IKKβ specific Abs and protein A-coated beads. The protein levels of ADAR1 p110 and P150 were detected using WB analysis with anti-ADAR1 specific Abs. Rabbit IgG served as negative control. Total cell lysate (input), and IP (bound) fractions served as positive control. (D) Representative WB analysis of ADAR1 p110 and p150 isoforms and IκB protein level in untreated, siControl and siADAR1 transfected HepG2 cells. TNF treatment served as positive control for NFκB pathway activation.
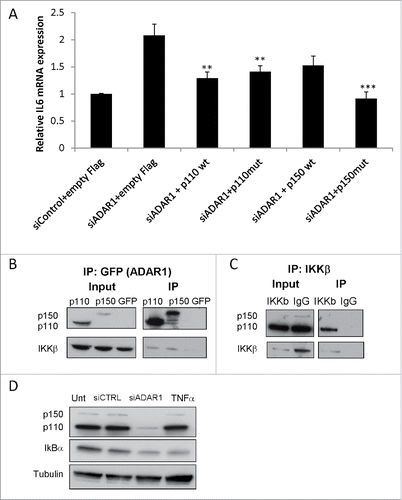
Figure 7. IL6 secreted from ADAR1 HepG2-KD cells mediates paracrine stellate cells activation. (A) Primary hepatic stellate cells were incubated with conditioned medium collected from siControl, siADAR, siIL6 or siADAR1and siIL6 co-transfected HepG2 cells. Relative mRNA expression levels of αSMA and Col1A in the HSCs were determined by qPCR analysis normalized to HPRT. Bar graphs show mean ± SEM of n = 3 experiments. (B) Representative microscopic images of the phenotypic changes of hepatic stellate cells following incubation with conditioned medium of siControl, siADAR1, siIL6, siADAR1+siIL6 transfected HepG2 cells (inverted light microscopy, x10 magnification).
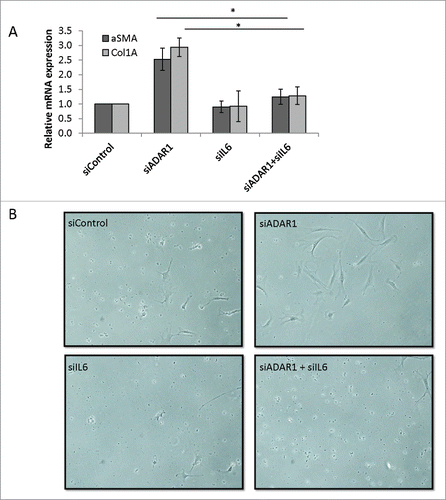
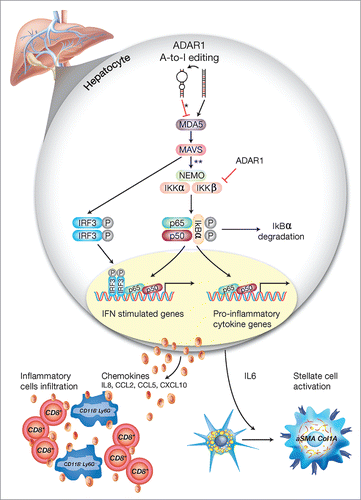
Figure 8. A schematic model of the regulatory role of ADAR1 in liver homeostasis. Under homeostatic conditions, ADAR1 catalyzes the promiscuous A-to-I editing of duplex-forming repetitive elements within mRNAs to reduce their potential to serve as ligands for the cytosolic dsRNA sensor, MDA5 (* refer to Citation33). In addition, as we now show, ADAR1 interacts with IKKβ to regulate the activity of the canonical NFκB pathway. Thus, lack of ADAR1 in hepatocyte induces aberrant signaling downstream of MDA5 leading to MAVS-dependent activation of IRF3 and NFκB transcriptional activity (** refer to. Citation13,16,18). Low ADAR1 levels are strongly associated with enhanced degradation of IκBα and the nuclear translocation of p65/p50 dimers to induce IL6 transcription. Moreover, the reciprocal activation/nuclear translocation of IRF3 promotes transcription of type-I interferons and multiple ISGs. This aberrant synergistic activation of the NFκB and type-I interferons pathways in hepatocytes, upon ADAR1 depletion, results in IL6-dependent paracrine activation of stellate cells to display a fibrogenic phenotype (Col1A+/aSMA+) alongside the recruitment of CD8+ cytotoxic lymphocytes and CD11B+ Ly6G+ granulocytes, further mediating multifocal liver damage and fibrosis.