Figures & data
Figure 1. Models of U2AF2-dependent and structure-dependent splicing. (A) U2AF2 recognizes the polypyrimidine tract upstream of the 3ss. It dimerizes with U2AF1 (not shown) which recognizes the 3ss AG to facilitate U2 docking on the 3ss. The interactions between U1 and U2 snRNPs bring the 5ss and 3ss in close proximity, forming a pre-splicing complex (A complex) toward the first step of splicing. (B) A special class of introns identified in fish and lamprey contains complementary runs of AC's and GU's across introns. The complementarity bridges the introns so that the 5ss and 3ss are positioned for splicing. (C) Intronic bridging by complementary repeat sequences were also suggested in humans. A variety of G, C and GC-rich repeats could play structural roles in pairing splice sites across introns. (D) Highly polymorphic genes, such as human HLAs exhibit rapid and frequent sequence changes in the coding exons to accommodate the need of peptide variety. Therefore the exonic splicing elements, for example exonic splicing enhancers (ESE's), are not preserved. Introns flanking the highly polymorphic exons are GC-rich and have lower folding energy compared to introns within the same length range. We hypothesize that the secondary structures of such introns bridge the splice sites, similarly to the fish AC- and GT-repeat containing introns, which compensate for the potential loss of exon definition. Red lines in the exons indicate single nucleotide polymorphisms.
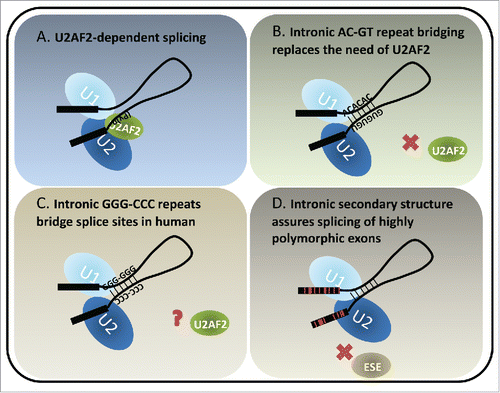
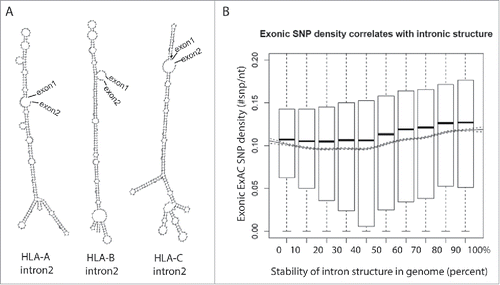
Figure 2. Highly polymorphic exons are flanked by structured introns. (A) Structures of intron 2 of the HLA family genes are predicted using an RNA structure prediction program, RNAfold.68 (B) Introns are ranked in decile by their folding energy calculated by RNAfold. SNP density of their flanking exons is calculated using all SNPs from the Exome Aggregation Consortium (ExAC) data and presented by boxplot for each bin. Trend of means (solid line) is depicted with 95% confidence interval (dashed lines). The significance of the positive correlation is determined by trend test, P value < 2 × 10−16.