Figures & data
Figure 1. Tritium incorporation assay suggests that Dnmt2 methylates a tRNA containing a deoxycytidine at position 38. (A) Cloverleaf structure of unmodified human cytosolic tRNAAsp. Position 38 has been engineered to contain rC, dC, or dm5C in three hybrid tRNAs of corresponding name. (B) In vitro methylation measured by tritium incorporation from 3H-SAM. Standard deviations of three replicates are given in Fig. S1.
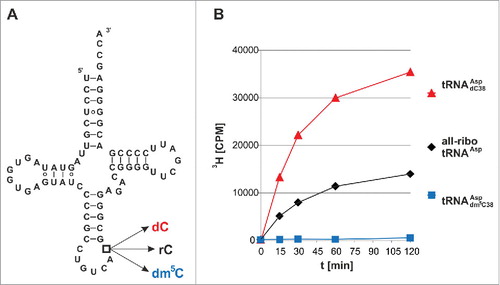
Figure 2. LC-MS analysis confirms that Dnmt2 methylates a tRNA containing a deoxycytidine at position 38. (A) Fragmentation patterns and mass transitions used for scanning of rm5C and dm5C in LC-MS. (B) LC-MS analysis of all-ribo tRNAAsp and hybrid tRNAAspdC38 before and after in vitro methylation by human Dnmt2.
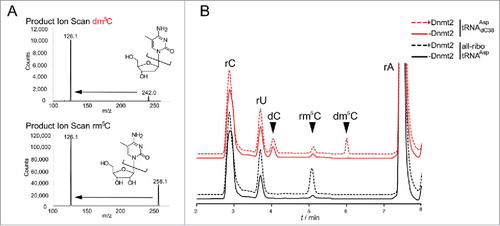
Figure 3. Dnmt2 methylates hybrid tRNAs containing up to 10 deoxynucleotides. (A) Positions of DNA substitutions in the tRNA cloverleaf in hybrid tRNAs. (B) Methylation of hybrid tRNAs of increasing DNA content. Average values and standard deviations are given in Fig. S1.
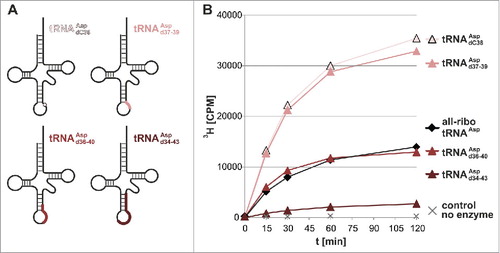
Table 1. tRNA-derivatives constructed by ligation and/or hybridization. Numbering in column 2 corresponds to that used in .
Figure 4. Relative methylation efficiency of covalent chimeric tRNAsAsp compared to the all-ribo tRNAAsp (entry 4c in ), which is set to 100 on the black scale bar on the right.
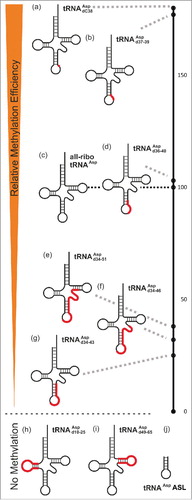
Figure 5. Relative methylation efficiency of 2-component hybrid-chimeric tRNAsAsp compared to the all-ribo tRNAAsp (entry 4c/5b in ) which is set to 100 on the black scale bar in the middle. Deviations from the native tRNAAsp sequence are presented in capital letters, DNA is highlighted in red.
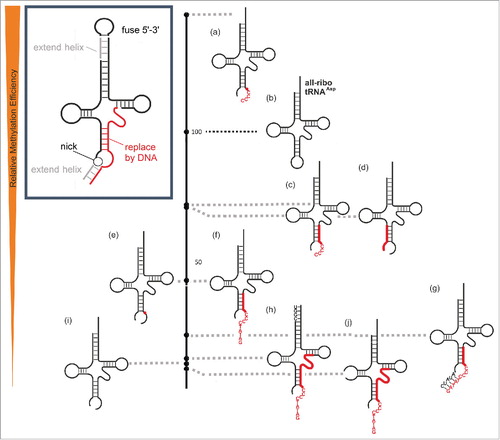
Figure 6. Methylation of an RNA-guided DNA oligonucleotide by Dnmt2. (A) Structure of the hybridized construct. DNA shown in red. (B) Average values and standard deviations of 3 tritium incorporation assays are shown.
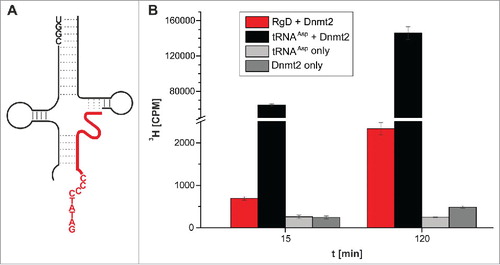
Figure 7. (A) Two-dimensional thin-layer chromatography of nucleosides on a 10 cm x 10 cm cellulose TLC plate. The starting point is marked by an X. (B) Methylation of an RNA-guided DNA oligonucleotide by Dnmt2. The oligonucleotides were hydrolyzed to nucleosides after the tritium incorporation assay, separated by 2D thin-layer chromatography and analyzed with the Cherenkov counter. Tritium could only be detected in m5dC. Average values and standard deviations of 3 experiments are shown. Control corresponds to background signal of the TLC plate. Note that an identical Fig. S4 with enhanced contrast can be found in the supplement.
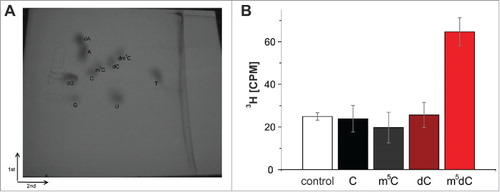
Table 2. Oligonucleotides used in this study. Ribonucleotides are indicated by plain letters, deoxyribonucleotides are indicated as dC, dT, dA, dG, or dm5C.
Table 3. Mass transitions for MRM and retention times in LC-MS analysis.
