Figures & data
Figure 1. KOD1Rnl purification and predicted structure. (A) Bioanalyzer protein electrophoresis gel plot of crude and purified KOD1Rnl. (B) Protein eletropherogram of crude and purified KOD1Rnl. (C) Circular dichroism spectra of KOD1Rnl before and after thermal denaturation. (D) The CD thermal denaturation curve of KOD1Rnl at 210 nm. (E) Predicted 3D structure of KOD1Rnl from both homology and ab initio based protein folding.
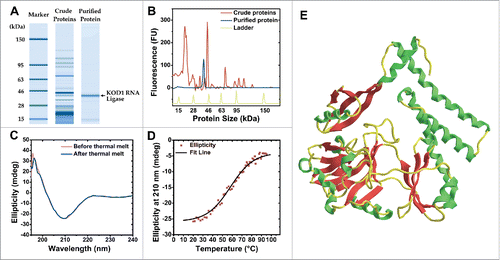
Figure 2. Alignment and structure of KOD1Rnl motifs. (A) Protein sequence of KOD1Rnl aligned against that of T4Rnl2, showing the conserved motifs (I-V) of Rnl2-like ligases. The five motifs are highlighted in yellow. Fully or semi- conserved amino acids within the motifs are bolded. (B) The predicted structures of the 5 motifs in KOD1Rnl. The β sheets are displayed in red, and the random coils are displayed in yellow. The amino acid residues are labeled.
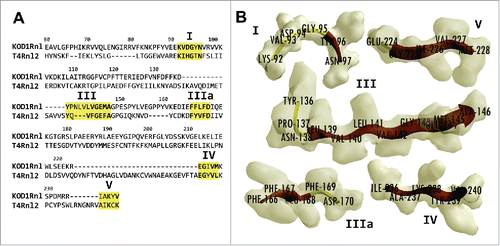
Figure 3. RNA detection using KOD1Rnl. (A) Illustration of RNA detection using thermostable double-stranded RNA ligase. (B) The effects of the terminal nucleotide on ligation probes, and the addition of RecJ, on both the template dependent and template independent ligation efficiencies. Reactions with or without RecJ were left for an hour at room temperature. (C) The effects of the time of incubation at 37 °C in the presence of RecJ, on the template dependent and template independent ligation efficiencies (*not attempted). The ligation probes used terminates in 2′ O-Methyl RNA analogs on the 3′ end. (D) The effects of different mismatched ligation probe terminal nucleotides on ligation efficiency. (E) Detection of different concentrations of Ebola RNA. (F) Illustration of the ligation probe hybridization regions on the Ebola RNA transcripts, and the corresponding primer binding regions on the joined ligation probes. Error bars denote 1 s.d.
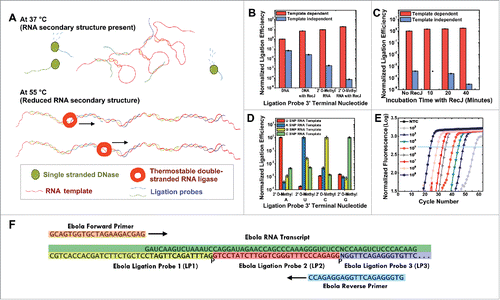
Figure 4. The relationship between the ratio of the dimensionless velocities and the ratio of the initial substrate concentrations. The ratio of the dimensionless template dependent velocity to the dimensionless template independent ligation velocity is plotted against the ratio of the concentrations of the initial dependent substrate to that of the initial independent substrate. The relationships are evaluated for different regimes of α and β. The hashed lines denote when the ratio of the dimensionless velocity is directly proportional to the ratio of initial substrate concentrations.
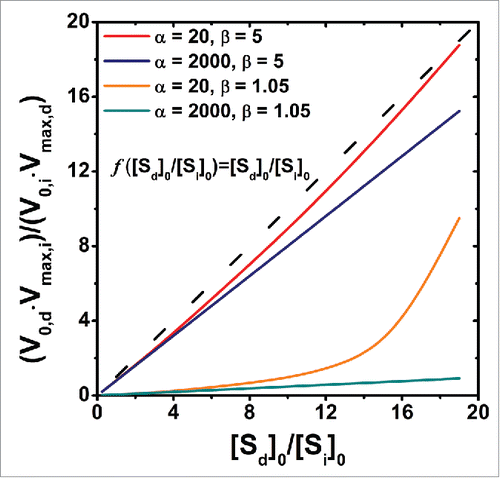
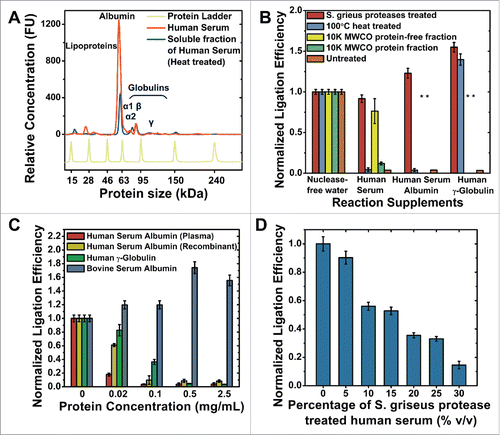
Figure 5. The effects of serum proteins on KOD1Rnl. (A) Bioanalyzer protein electropherogram of unprocessed human serum, and the soluble fraction of human serum after heat treatment. (B) Template dependent ligation efficiency of KOD1Rnl in the presence of different treated human serum, serum albumin and gamma-globulin. (C) Ligation efficiency of KOD1Rnl in the presence of different concentrations of plasma-derived and recombinant human serum albumin, human gamma-globulin, and bovine serum albumin. (D) The effects of the addition of different concentrations of S. griseus protease treated human serum on ligation efficiency. Error bars denote 1 s.d.