Figures & data
Figure 1. Contact of the 5′ region of mRNA CDS with the solvent side of 40S ribosome and eIFs in a 48S complex. (A) Experimental outline of cross-linking assays using a minimal version of Sindbis virus (SV) mRNA bearing photoactivatable 4-thio UTP residues at the indicated positions in the DLP structure (rounded dots, SV-U1; squared dots, SV-U2). [32P]-labeled mRNAs were used for the assembly of 48S complexes in RRL in the presence of GMP-PNP as described recently.Citation26 Ribosomal fraction was purified by sedimentation through a sucrose cushion at 200,000xg for 3h and analyzed. (B) An aliquot of the sample was used to analyze mRNA-18S rRNA cross-linking by primer extension with RT as described previously.Citation26 The presence of extension arrests corresponding to the ES6S region of 18S rRNA is indicated. The rest of the sample was digested with RNAse A and U1, and the resulting [32P]-cross-linked proteins were analyzed by SDS-PAGE followed by autoradiography. (C) Protein bands detected in 48S complex or in 80S complex assembled in the presence of GMP-PNP and cycloheximide, respectively. (D) The identity of a 48 kDa band was confirmed by immunoprecipitation under denaturing conditionsCitation32 using an anti-eIF4A antibody (St. Johns, STJ27247) or a control antiserum against RPS2 (Santa Cruz Biotech. ). (E) The eIF4A protein band was further confirmed by treatment with hippuristanol, a specific inhibitor that prevents the RNA-binding activity of eIF4A.Citation28 The intensity of p52 band greatly varied among experiments, depending on the RRL batch used and the ultracentrifugation conditions. The molecular weight markers are shown. WRF: whole ribosomal fraction. is adapted from Toribio et al. (2016).Citation26
![Figure 1. Contact of the 5′ region of mRNA CDS with the solvent side of 40S ribosome and eIFs in a 48S complex. (A) Experimental outline of cross-linking assays using a minimal version of Sindbis virus (SV) mRNA bearing photoactivatable 4-thio UTP residues at the indicated positions in the DLP structure (rounded dots, SV-U1; squared dots, SV-U2). [32P]-labeled mRNAs were used for the assembly of 48S complexes in RRL in the presence of GMP-PNP as described recently.Citation26 Ribosomal fraction was purified by sedimentation through a sucrose cushion at 200,000xg for 3h and analyzed. (B) An aliquot of the sample was used to analyze mRNA-18S rRNA cross-linking by primer extension with RT as described previously.Citation26 The presence of extension arrests corresponding to the ES6S region of 18S rRNA is indicated. The rest of the sample was digested with RNAse A and U1, and the resulting [32P]-cross-linked proteins were analyzed by SDS-PAGE followed by autoradiography. (C) Protein bands detected in 48S complex or in 80S complex assembled in the presence of GMP-PNP and cycloheximide, respectively. (D) The identity of a 48 kDa band was confirmed by immunoprecipitation under denaturing conditionsCitation32 using an anti-eIF4A antibody (St. Johns, STJ27247) or a control antiserum against RPS2 (Santa Cruz Biotech. ). (E) The eIF4A protein band was further confirmed by treatment with hippuristanol, a specific inhibitor that prevents the RNA-binding activity of eIF4A.Citation28 The intensity of p52 band greatly varied among experiments, depending on the RRL batch used and the ultracentrifugation conditions. The molecular weight markers are shown. WRF: whole ribosomal fraction. Fig. 1B is adapted from Toribio et al. (2016).Citation26](/cms/asset/34efbe77-827d-4cc7-926a-c37033998c22/krnb_a_1247146_f0001_b.gif)
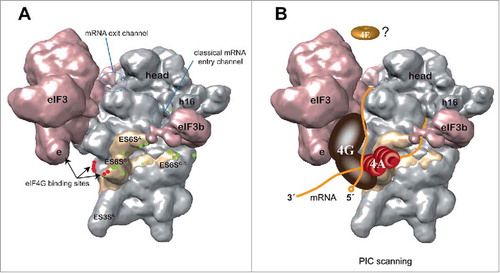
Figure 2. Model of PIC scanning based on data from an alphavirus mRNA 48S complex. (A) Solvent side view of a 43S PIC model (EMD-5658)Citation16 showing the cross-linking sites (green dots) of alphavirus mRNA DLP with the ES6S region of 18S rRNA (pale yellow). The data are derived from experiments such as those shown in . The ES6S region is made up of 3 main RNA stems as indicated. The binding sites of eIF4G to the eIF3e subunitCitation6 and ES6 helices (red dots)Citation27 are also indicated. The entry and exit regions define the minimal channel where mRNA is threaded for decoding (60S subunit side, not visible). (B) A model of PIC scanning that integrates most data published to date. According to this model, mRNA enters the ribosome through (or near) the ES6S region where eIF4A bound to eIF4G may be placed, interacting with the incoming region (3′) of mRNA for secondary structure unwinding. The elongated structure of scaffold eIF4G may span from the right arm of eIF3 (e subunit) to ES6S, 2 regions that make contact with the middle domain of eIF4G.Citation17 The probable dissociation of eIF4E from the cap structure of mRNA upon 48S assembly is based on recent data.Citation13