Figures & data
Figure 1. Regulation of TF binding by promoter-associated ncRNAs in fission yeast. (A) Schematic diagram of the fbp1+ locus. Each number represents a location relative to the start site of the fbp1+ open reading frame. UAS1 and UAS2 are cis-acting elements involved in transcriptional activation of fbp1+.Citation16 (B) Models for how promoter-associated ncRNAs (including mlonRNAs) enhance Atf1 binding. (i) Groucho/Tup1-like corepressors Tup11 and Tup12 (represented by “Tup” in the figure for simplification) repress Atf1-DNA association, and this inhibition is locally attenuated by ncRNAs transcribed near Atf1-binding sites. (ii) ncRNAs can also facilitate Atf1 binding independently of Tup11/12.
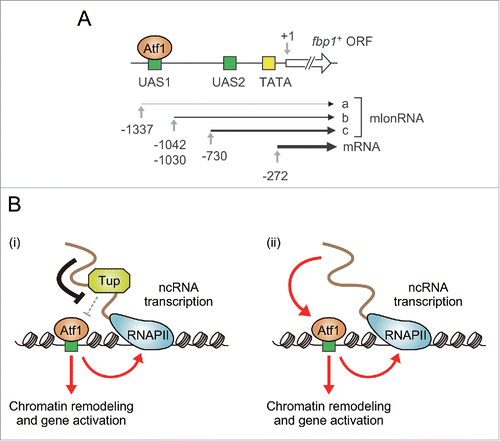
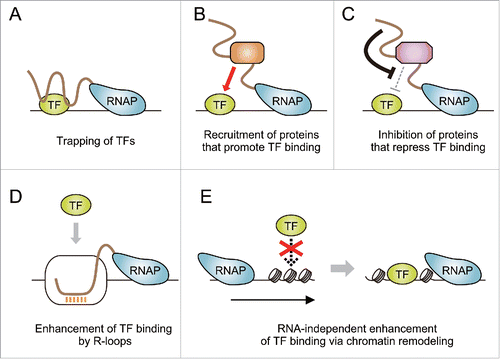
Figure 2. Possible models for how TF binding is driven by on-site transcription of ncRNAs. (A) Nascent ncRNAs trap TFs at their target DNA regions. (B) ncRNAs recruit proteins that assist TF binding (e.g., histone modifiers and chromatin remodelers that create open chromatin structure). (C) ncRNAs attenuate functions of proteins that play inhibitory roles for TF binding (e.g., corepressors that establish chromatin states refractory to TF binding). (D) ncRNA transcription leads to the formation of R-loops that facilitate TF binding. (E) Transcription-coupled chromatin reorganization promotes TF binding independently of RNA products.