Figures & data
Figure 1. RipSeq protocol retains BicD/Egl complex stability and enriches for known target mRNAs. (A-B) Extracts from Egl::GFP expressing embryos and wt control extracts were subjected to IP with anti-GFP antibodies. Western blotting tested for the presence of the BicD and Egl::GFP (A). Semiquantitative RT-PCR tested for the enrichment of known BicD/Egl mRNA targets and for the lack of enrichment of non-localizing mRNAs and house keeping mRNAs (B). (C) Rip-Seq enrichment of mRNAs from the test set. (D-E) Most top candidate BicD/Egl targets identified by Rip-Seq (D) could be validated by RT-qPCR analysis in an independent IP experiment (E). Error bars represent +/− SD of 2 independent IPs. Note that fold enrichment values of the 2 different experiments cannot be compared directly because they are calculated using different formulas (see methods). Nevertheless, enrichments in the Egl::GFP IPs relative to mock IPs is reproducible.
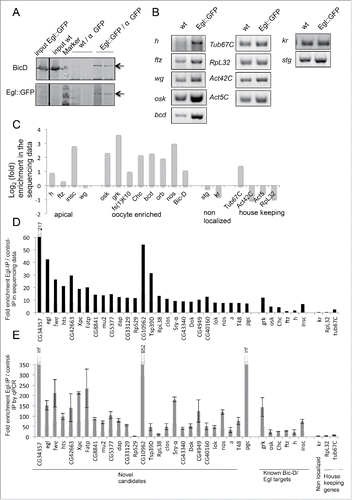
Table 1. Localization patterns of Egl::GFP target mRNA candidates in embryonic stages 1–5.
Table 2. Localization patterns of Egl::GFP target mRNA candidates in ovaries.
Figure 2. Egl targets are enriched for mRNAs localizing to pole plasm and pole cells. 21% (10/47) of the BicD/Egl targets localize to the pole plasm or pole cells compared to 8.4% of a random set.Citation6 (A) Egl protein is enriched in the pole plasm (upper panels, embryo in syncytial division before polar bud formation) and in pole cells (lower panels, embryo in late stage 4). (B) BicD protein signal is detected slightly enriched in pole plasm and pole cells. Stages are as in A). (C) In situ hybridization to whole-mount wild-type embryos (wt, OreR) using antisense RNA probes for the candidate mRNAs CG8841, lok, Dok and CG33129. Hoechst (blue) visualizes the DNA. Scale bars are 30 μm.
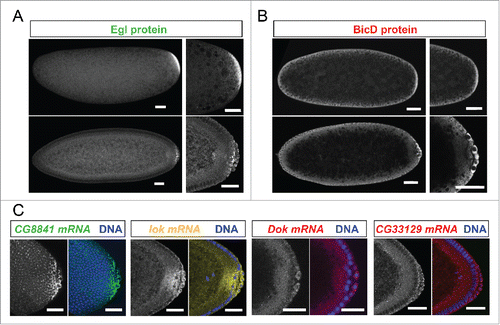
Figure 3. Egl targets show specific and novel apical localization patterns. Apically localizing mRNAs are 6-fold enriched compared to a published random set (10/47; 21% vs 3.4%Citation6). In situ hybridizations to wild-type (OreR) embryos oriented anterior to the left and dorsal up. Hoechst (blue) visualizes the DNA. (A) Novel transcripts showing continuous apical localization in blastoderm embryos (red signal). Apical localization varies and changes for different mRNAs and nuclear cycles, respectively. RpL38 mRNA in situ was used as a control that shows the distribution of a non-localizing mRNA. (B-D) Novel candidates expressed apically but not continuously from anterior to posterior. (B) arc (a) is enriched apically of the nuclear layer in the anterior region of the embryo (green signal). (C) T48 is enriched apically only in the ventral region (green signal). (D) CG8841 shows apical enrichments at the anterior and posterior poles (including pole cell expression) and in the middle of the embryo (green signal). CG33129 (red signal) shows apical localization in the invaginating furrows during gastrulation (E) and in tracheal precursor cells (stained for the Tango (Tg) marker in green) later in embryogenesis (F). Scale bars represent 25 µm except in (F) were they represent 30 µm. Histograms depicted on the right side of the pictures show the fluorescence intensity along the apical-basal axis of the marked region around a blastoderm nucleus (dotted square). This region is also shown magnified on the right side.
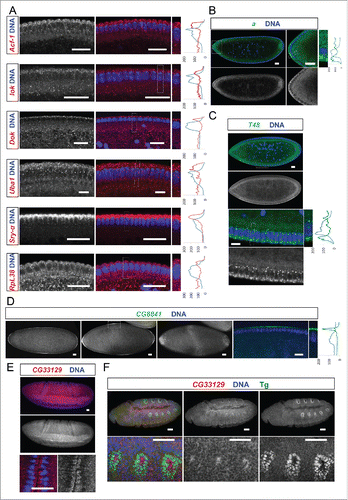
Figure 4. Egl targets are enriched for mRNAs that localize to the oocyte in a BicD dependent manner. 61% of the BicD/Egl targets tested showed accumulation in the oocyte compared to the expected 17% observed in a random set of mRNAs. In situ hybridization to wild-type (OreR) controls and to egg chambers 4 d after turning off BicD expression (BicDmom). Antisense RNA probes for the candidate mRNAs were labeled with green or red fluorescent signals. Almost no BicD mRNA signal is observed when BicD expression is off (green signal, upper-most panels). During early wild-type oogenesis, clos, fwe, lok, CG43340, nos, T48, and a accumulated in the oocyte where they became enriched at the posterior till stage 6. Subsequently the signal appeared at the anterior cortex by stage 7, showing this pattern until mid to late oogenesis (left panels). These mRNAs failed to efficiently accumulate in BicDmom oocytes when BicD was off and the late localization in the oocyte was also severely impaired in BicDmom oocytes (right panels). CG6459 showed expression in the oocyte cytoplasm at all stages. However, in this case the oocyte signal was not enriched compared to its accumulation in nurse cells. A weak BicD dependent concentration of the mRNA at the dorsal side of the oocyte nucleus was observed by stage 8 (arrowhead, and magnified region in inset picture). By stage 9 a more clear presence of a dotted CG6459 signal was observed in the oocyte, but levels did not seem to exceed nurse cell levels. In BicDmom ovarioles big blobs of CG6459 mRNA signal were seen in the nurse cell cytoplasm, suggesting a problem in transport of this mRNA. Hoechst (blue) visualizes the DNA. Scale bars are 20 µm.
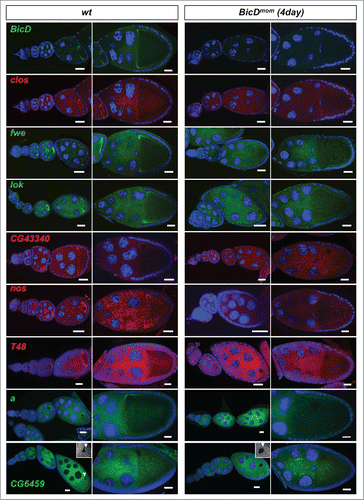
Figure 5. BicD is needed for apical mRNA localization in follicle cells. 6.5% of the mRNA tested show apical localization in follicle cells. This mRNAs class is enriched 8-fold among the Egl::GFP targets compared to a random set (6.5% vs 0.8%Citation11) (A) In situ hybridization to wild-type (OreR, wt) controls and to egg chambers 4 d after turning off BicD expression (BicDmom). Oocyte enrichment (arrows) as well as apical follicle cell localization of CG33129, egl and Uba1 was impaired when BicD was off. High magnification of the stage 10 follicle cell epithelium is shown in the right panels. Note that apical is toward the oocyte (up in the magnified pictures). Scale bars are 20 µm. Uba1 apical enrichment is weaker and less cortical, but still requires BicD. Histograms depicted on the right side of the pictures show the fluorescence intensity along the apical-basal axis of the marked cortical region around the nuclear layer (dotted square). This region is also shown magnified on the right side of the micrographs. (B) BicD mRNA expression (red) is drastically reduced in stage 9–10 BicDmom egg chambers in the germline and also in the somatic follicle cells (high power pictures on the right). Note that BicD mRNA is also apically localized as expected since it is also a target of Egl. (C) BicD protein (red) is also apically enriched in wild-type follicle cells, but is almost undetectable in BicDmom egg chambers, oocytes and follicle cells. Hoechst (blue) visualizes the DNA. Scale bars are 20 µm.
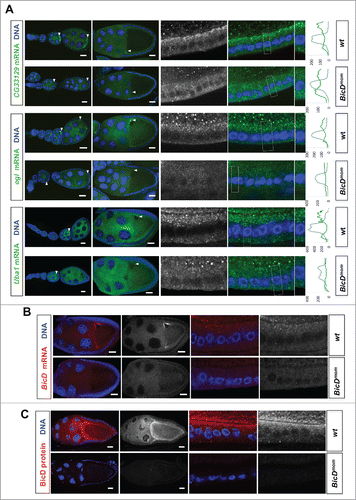
Figure 6. 3′ UTR hairpins of Dok and lok are responsible for apical localization. (A, E) Schematic representations of the test lok (A) and Dok (E) mRNAs injected. (B, F) Secondary structure of the localizing hairpins in lok (B) and Dok (F) predicted using the RNAfold web server. (C, D, G) Representative images of embryos injected with the respective lok (C-D) and Dok (G) constructs indicated above each picture. Fluorescently labeled transcripts (green signal) were injected into blastoderm embryos basally and fixed 12 min thereafter. Images are oriented with apical at the top. Hoechst (blue) visualizes the DNA. The categorization of the RNA enrichment to the apical cytoplasm for the corresponding injections shown in (C, D, G) is shown under each embryo image (C', D', G'). The percentage of embryos showing strong (++), weak (+), very weak (+/−) or no apical (−) localization is shown for each construct. (N) Number of embryos scored for each mRNA injection. Scale bars are 10 µm.
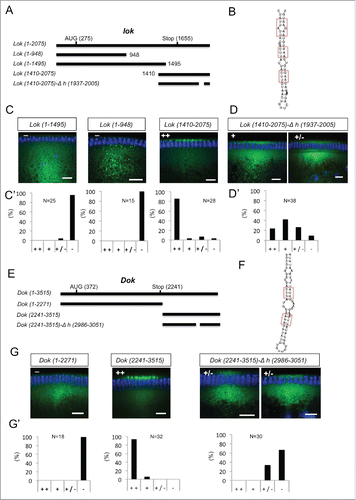
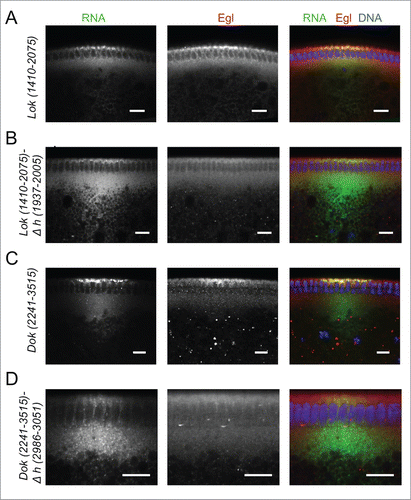
Figure 7. Recruitment of Egl to apically localizing Dok and lok 3′ UTRs. (A-D) Immunostaining revealed the distribution of Egl (red) following basal injection of lok (green) (A, B) and Dok RNA fragments (C, D) into wild-type embryos. Egl is co-recruited to apically localized Dok and lok 3′ UTR sequences (A, C), but not if the localizing hairpin sequence was deleted from the construct (B, D). Scale bars are 10 µm.