Figures & data
Table 1. Inventory of rRNA modifications in yeast. The rRNA, position (Posn.) and type (Mod.) of the modification and the enzyme/snoRNP(s) that introduce it are given, with information on whether partial modification was observed (✓; <85%)Citation21 or not (x), and whether modification occurs early/chromatin-associated (E),Citation18 in late nucleolar/nuclear (LN) particles or in the cytoplasm (C). A lack of information on the timing is indicated by “–.” Enzymes/snoRNPs targeting the same position sequentially are separated by “,” and alternative enzymes/snoRNPs that can install a single modification at a given site are separated by “/”. The information presented in this table is compiled from various references cited in the text.
Figure 1. Structures of box C/D and box H/ACA sRNP complexes. (A) Structural model of an H/ACA box sRNP from Pyrococcus furiosus (PDB 3HAY)Citation154 A surface view of the protein components is shown and sRNA and rRNA are indicated in dark gray and red, respectively. (B) Structural model of a Sulfolobus solfataricus box C/D sRNP (PDB 3PLA)Citation155 is shown as in A. The modified nucleotide is shown in surface view. Note, that archaeal L7Ae is an ortholog of eukaryotic Snu13 and in eukaryotes, Nop56 and Nop58 are orthologous to archaeal Nop5.
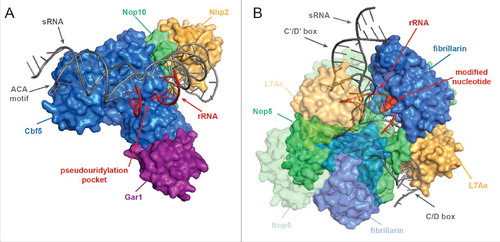
Figure 2. Base modifications in rRNA and the enzymes that install them. (A) Chemical structures of the 4 nucleotides and the modifications that are added in yeast rRNAs. The additional chemical groups are marked in red and the enzymes that introduce them in yeast are indicated above the arrow (yellow) and in humans below the arrow (green). (B) Three-step modification pathway for U1191 of the 18S rRNA in yeast.
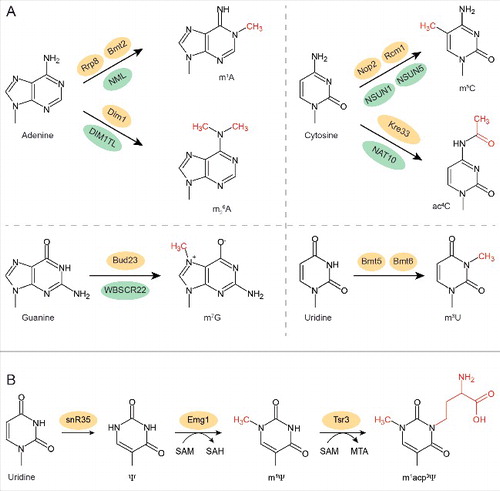
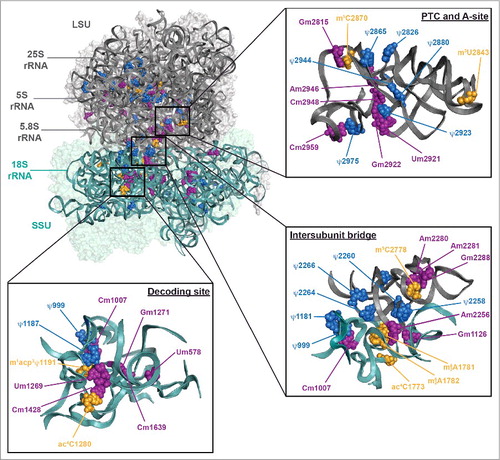
Figure 3. Distribution of rRNA modifications on the yeast ribosome. The S. cerevisiae 80S ribosome (PBD 4V88)Citation6 is shown – 40S in teal and 60S in gray. The positions of 2′-O-methylations (purple), pseudouridines (blue) and base modification installed by stand-alone enzymes (orange) are indicated. Three functionally important regions of the ribosome, the peptidyltransferase center (PTC), the decoding site and the intersubunit bridge eB14, are also shown in a magnified view.