Figures & data
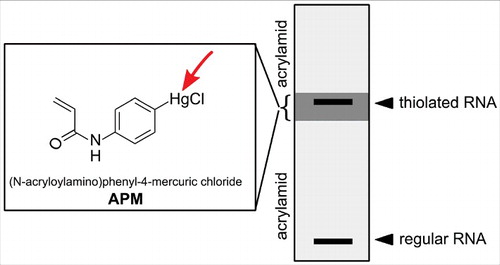
Figure 1. Overview of modified nucleosides. The arrows indicate the target sites in the modified nucleosides responsible for their reactivity with the described reagents. Red arrows: covalent reaction, green arrows: converting reaction, blue arrows: physicochemical interaction.
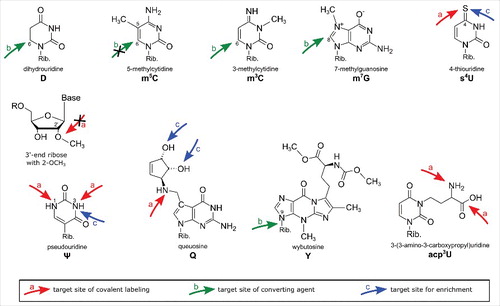
Table 1. Overview of all reagents used for covalent labeling and detection of modified nucleosides. Red arrows indicate the active groups responsible for the reaction with the target modification. The term “target modification” lists the modified nucleosides known to react with the reagent from the literature. “Potential side reactions” lists all nucleosides that are chemically able to react with the reagent but were not described in the literature.
Table 2. Overview of conversion products after bisulfite reaction of cytidine and its modified derivatives. Without pretreatment (2nd column from left) C, f5C and ca5C convert into uracil after bisulfite treatment. Catalytic oxidation of hm5C into f5C (3rd column from the left) leads to its conversion into uracil after bisulfite treatment while all cytidines are protected from conversion after enzymatic methylation (first column from the right). Thus f5C and ca5C become detectable.
Figure 2. Reaction of carbodiimides with pseudouridine and uridine. Top: acylation of pseudouridine (Ψ) with the carbodiimide CMCT (full name in text). The carbodiimide group reacts with both N1 and N3, but after alkaline treatment the N1-CMCT is cleaved. The remaining CMC at N3 enables detection by mass spectrometry, reverse transcription, sequencing or even biotin pulldown in case of the CMC-azide (encircled, blue). Bottom panel: Like pseudouridine, uridine gets labeled at the N3 position, which is removed after alkaline treatment. The same was found to be true for guanosine and inosine. Insert: Structure of CMCT.
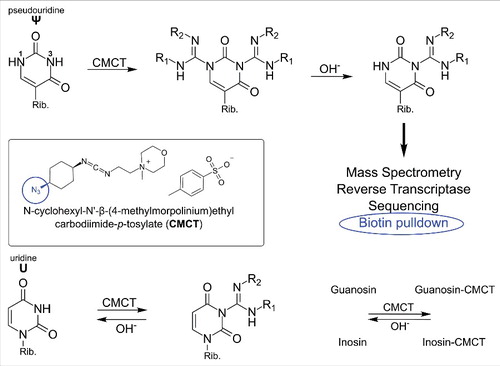
Figure 3. Conversion of different modified nucleobases and reagents. (a) Mechanism of cytosine to uracil conversion by bisulfite treatment. 5-methylated cytosine (m5C) does not react with bisulfite, due to its lower electrophilicity, and thus is not converted to uracil. (b) Strand break analysis reveals the presence of modified nucleosides after treatment with various reagents. These reactions with modified bases lead to e.g. depurination which results in strand breaks after anilin treatment. In contrast, 2′O-methylated ribose is protected from alkaline induced cleavage which allows detection of the methylated site.
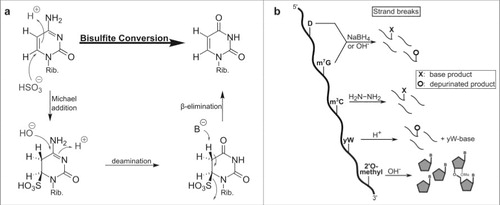
Figure 4. Enrichment of thiolated RNA on a 3 layered acrylamide gel. The upper and lower gel layer allow normal migration of RNA, while the 2nd layer contains APM (structure shown in the box) which retains all thiolated RNA.