Figures & data
Figure 1. Deazaguanine derivative synthesis pathways. GTP is the preQ0 precursor in both bacteria and archaea (purple arrows). In most bacteria, four more enzymatic steps lead to the insertion of Q in tRNAs at position 34 (red arrows). In a few organisms, preQ0 can be transformed to secondary metabolites such as toyacamycin or sangivamycin antibiotics (red arrows, toy genes). In eukaryotes, queuine is salvaged (green circle) and directly transferred to tRNAs (green arrows). Bacteria salvage preQ1 (red circle), and both bacteria and archaea salvage preQ0 (purple circle). In archaea, preQ0 is transferred to position 15 of tRNA before being modified to G+ (blue arrows). PreQ0 and ADG have been found in bacterial DNA (dashed red arrows) and G+ in phage DNA (dashed yellow arrow). All dashed arrows represent uncharacterized reactions. All molecule abbreviations and protein names are described in the text.
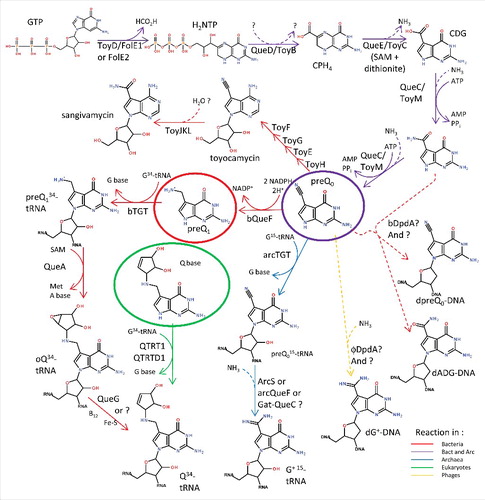
Figure 2. Catalytic and substrate recognition residues in different TGT subgroups. Partial alignments of Zymomonas mobilis (Zm) bTGT (PDB 1WKF), Escherichia coli (Ec) bTGT (RefSeq AMK98755), Homo sapiens (Hm) QTRT1 (RefSeq AAH15350), Homo sapiens QTRTD1 (RefSeq EAW79613), Pyrococcus horikoshii (Ph) arcTGT (PDB 1IQ8), Salmonella enterica serovar Montevideo (Sm) bDpdA (Genbank EFY12575), and E. coli phage 9g pDpdA (RefSeq YP_009032326). The conserved residue for substrate recognition, zinc binding, and catalytic activity are shown in bold and numbered. The arcTGT exhibits three supplementary C-terminal domains (C1, C2 and PUA), represented in black on the archaea line.

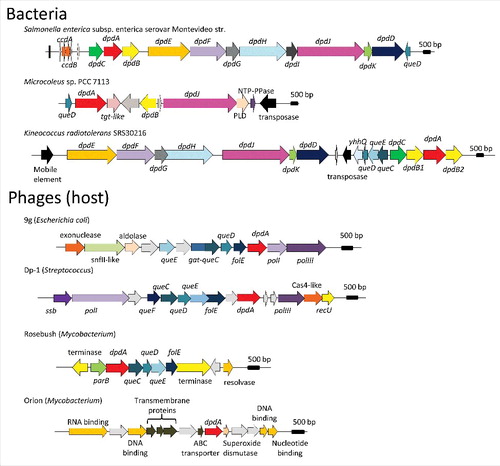
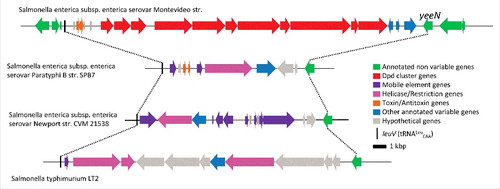
Figure 3. Genome neighborhoods of dpdA in bacteria and phages. The dpd and the queuosine related genes are defined in the text, general family memberships were noted for the other genes. The host of each phage is indicated between parenthesis aside of each phage name. Accession numbers for the corresponding DpdA proteins are: WP_001542917.1 for Salmonella enterica subsp. enterica serovar Montevideo str., WP_015211588 for Microcoleus sp PCC 7113, ABS05840 for Kineococcus radiotolerans SRS30216, YP_009032326 for Escherichia coli phage 9g, YP_004306895 for Streptococcus phage Dp-1, NP_817763 for Mycobacterium phage Rosebush, and YP_655116 for Mycobacterium phage Orion.