Figures & data
Figure 1. Anticodon loop modifications of tRNALysUUU and translational +1 frameshift models. (A) Schematic representation of tRNALysUUU anticodon loop with ct6A (6) and mcm5s2U (*) modifications. Gene-products relevant for synthesis of the modifications and addressed in this study are indicated. (B) Schematic representation of the modified Ty1 frameshift construct sensitive to tRNALysUUU modification defects.Citation28 Shown is the W12 construct where lacZ is in the +1 frame with respect to HIS4A.Citation28 The shift site as well as amino acids specified and +1 and 0 frame UGA stop codons are indicated. (C) With fully modified tRNALysUUU frameshift rates are low and termination at the UGA downstream of the Lys codon occurs efficiently. (D) Frameshift induction by tRNALysUUU modification defects via an A-site effect. Hypomodified tRNALysUUU (absence of * and 6, stippled) inefficiently binds to the AAA codon while peptidyl-tRNALeuUAG occupies the P-site. This pausing allows the peptidy-tRNALeuUAG to shift to the +1 frame and the appearance of an AAC codon in the A site (read by tRNAAsnGUU). This effect would be suppressible by extra copy of tRNALysUUU.Citation28 (E) Frameshift induction by tRNALysUUU modification defects via a P-site effect. Hyopmodified tRNALysUUU (absence of * and 6) binds to the AAA codon and translocation occurs. Once present in the P-site as peptidyl tRNALysUUU, binding is weakened (loosing P-site grip) and +1 frameshift occurs. This effect would not be suppressible by extra copy of tRNALysUUU.
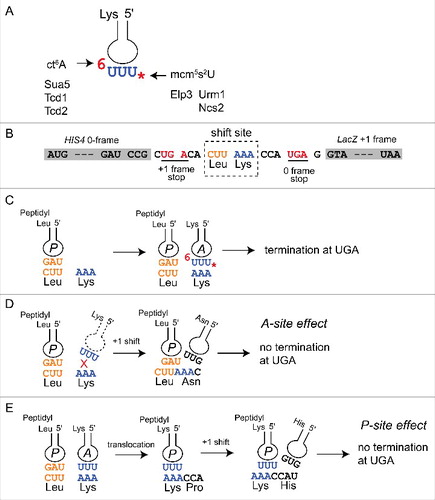
Figure 2. Induction of +1 translational frameshift rates relative to the wild type. Fold induction rates were calculated based on normalized frameshift values [%] shown in and significance of differences determined using two-tail t-test. n.s.: not significantly different. Heat indicates moderate thermal stress condition during growth (37°C) and + tK(UUU) refers to frameshift levels measured with the WA-2 and FA-1 constructsCitation28 that carry an additional copy of the tK(UUU) gene.
![Figure 2. Induction of +1 translational frameshift rates relative to the wild type. Fold induction rates were calculated based on normalized frameshift values [%] shown in Table 1 and significance of differences determined using two-tail t-test. n.s.: not significantly different. Heat indicates moderate thermal stress condition during growth (37°C) and + tK(UUU) refers to frameshift levels measured with the WA-2 and FA-1 constructsCitation28 that carry an additional copy of the tK(UUU) gene.](/cms/asset/c658530b-7f29-4d63-8e5b-92da53b12fe1/krnb_a_1267098_f0002_b.gif)
Table 1. Normalized frameshift rates in different yeast strains and growth conditions.
Figure 3. Effect of temperature shift on Ahp1 urmylation and Urm1 levels. (A) Sulfur transfer from cysteine to Urm1 results in formation of Urm1 thiocarboxylate, which is required for covalent attachment of Urm1 to Ahp1 (urmylation) and tRNA thiolation. (B) Temperature shift assay. urm1 mutants expressing TAP-tagged Urm1Citation25 were cultivated at 30°C in YNB media to OD600nm = 1 after which one half of the culture was shifted to 37°C. At indicated time points, total protein extracts were prepared and subjected to anti-TAP Western analysis to detect free Urm1 as well as the slower migrating Ahp1-Urm1 conjugate. Previous studies with ahp1mutants confirmed the the slower migrating band to result from Ahp1 urmylation by the tagged Urm1 variant.Citation25,26 To verify equal loading, anti-Cdc19 antibodies were utilized. (C) As in (B) but with the culture part that remained at 30°C. (D) Signal intensities of slower migrating band in (B) and (C) were quantified and relative levels of Ahp1 urmylation at 30°C or 37°C were calculated by dividing signal intensity at time points 2.5h, 5h or 7.5h by the one of timepoint 0.
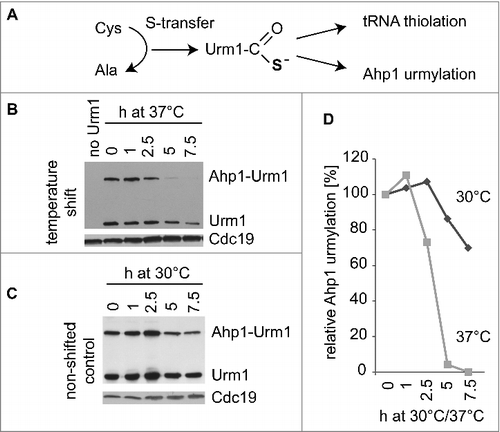
Figure 4. Heat induced growth defects in tcd1 and tcd2 single mutants are suppressible by elevated levels of tRNALysUUU. tcd1 and tcd2 mutants were transformed with either pRS425 (vector), pK (multi copy tRNALysUUU) or pQKE (multi copy tRNALysUUU, tRNAGlnUUG and tRNAGluUUC) and serial dilutions of cultures transferred to YPD plates that were incubated at the indicated temperatures for two days.
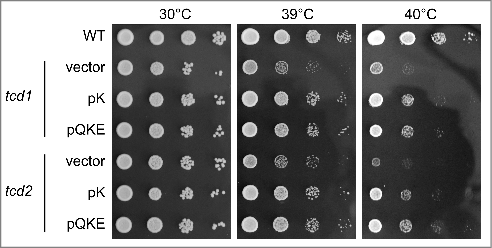
Table 2. Strains used in this study.
