Figures & data
Figure 1. Canonical tRNA structure with base 37 modifications. Canonical tRNA structure and 5 individual anticodon loops of tRNAs that carry different base 37 modifications. Where appropriate, base 32 and 34 modifications are indicated as well. Structural formulae indicate the modified nucleosides at position 37. Respective drawings of base modifications have been copied from www.modomics.genesilico.pl with permission.Citation14,15 Cm 2′O-methylcytosine; m1G 1-Methylguanosine; m3C 3-Methylcytosine, τm5U 5-taurinomethyluridine, Gm 2′O-methyl Guanosine, mcm5U 5-Methoxycarbonylmethyluridine, mcm5s2U 5-Methoxycarbonylmethyl-2-thiouridine, ms2i6A 2-Methylthio-N6-isopentenyladenosine, ms2t6A 2-Methylthio-N6-threonylcarbamoyladenosine, yW wybutosine.
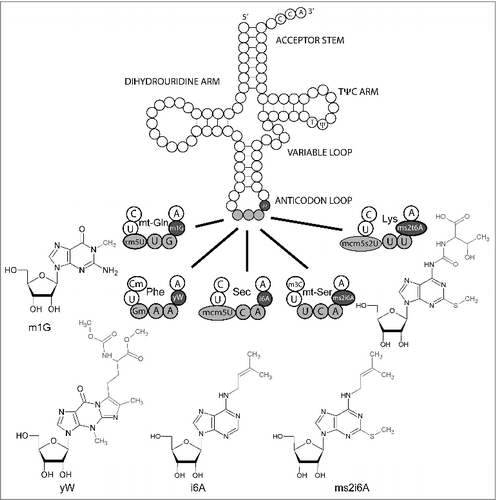
Table 1. E. coli tRNAs carrying i6A37 or ms2i6A37.
Table 2. Yeast tRNAs carrying i6A37 and tRNAs that carry i6A in other species.
Table 3. Mammalian tRNAs carrying i6A37 or ms2i6A37 and tRNAs that carry i6A in other species.
Figure 2. Effect of base 37 modification in tRNASec. (A) Unmodified murine tRNASec displays a disordered anticodon loop. Instead of forming a U-turn, U33 base pairs with unmodified A37 leading to an anticodon incapable of interaction with the codon.Citation78 (B) Model of fully modified tRNASec decoding the codon in the ribosome. The model was generated by the authors using MacPyMOL (Schrödinger) software and is based on PDB 2UUC containing a bacterial tRNAVal carrying 5-Oxoacetic acid-modified U34 and N6-Methyladenosine at position 37.Citation80
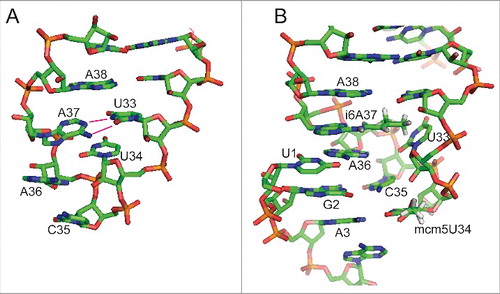
Table 4. Genes involved in tRNAs modification at position 37 in eukaryotes and associated phenotypes.
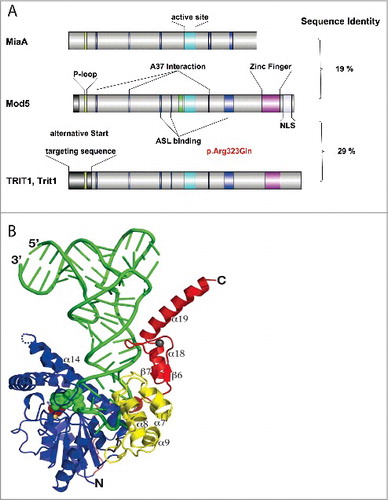
Figure 3. Structure of isopentenyl transferases and mode of substrate binding. (A) Comparison of isopentenyl transferases from E. coli (MiaA), baker's yeast (Mod5), and mammals (TRIT1, Trit1). Stretches of sequences with amino acid identity are highlighted in color. The sequence shown in green represents the P-loop involved in DMAPP binding. The turquoise sequence is the core of the active site. The blue sequences refer to amino acids interacting with the tRNA substrate (A37 binding or anticodon stem loop (ASL)-binding). The zinc-finger present in eukaryotes is depicted in violet. NLS, nuclear localization sequence. Overall sequence identity is shown on the right. The pathogenic mutation in human TRIT1 is indicated in red. The figure was prepared with IBS software.Citation95 (B) Crystal structure of Mod5- tRNACysGCA complex. The tRNA is shown in green. The DMAPP-binding and catalytic center in green balls, the zinc-finger domain is shown in red. Reproduced with permission from Zhou C and Huang RH. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: insight into tRNA recognition and reaction mechanism. Proc Natl Acad Sci U S A 2008; 105:16142–7.