Figures & data
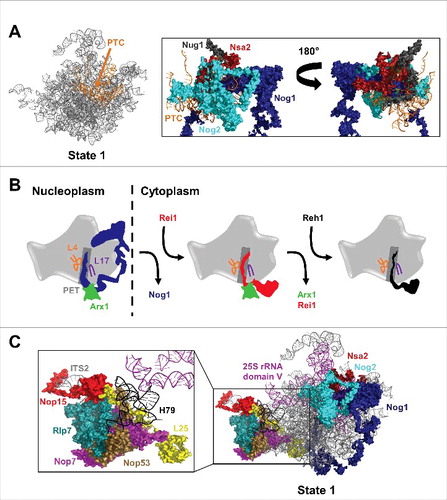
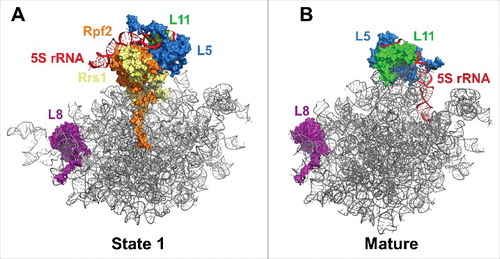
Figure 1. The yeast 60S subunit assembly pathway and AFs identified in state 1 and state 2 particles. (A) Timing of pre-rRNA processing events, entry and exit of AFs, and 5S RNP rotation. Also shown are the lifetimes of the Nog2-, Rix1-Rea1-, Arx1-, and Nmd3-bound pre-60S subunit assembly intermediates. The lifetimes of the state 1, 2, and 3 Nog2 particles are indicated by different shades of teal. The portions of the lifetimes of Arx1 and Nmd3 reported on by the Arx1 and Nmd3 particles are indicated by light green and gray, respectively. (B) The 19 AFs identified in the Nog2 state 1 particle (PDB ID 3JCT). The AF arc consists of 14 AFs stretching from the CP to the PET. Nop15, Rlp7, Nop53, Cic1, and Nop7 bind to ITS2. (C) The Nog2 state 2 particle.
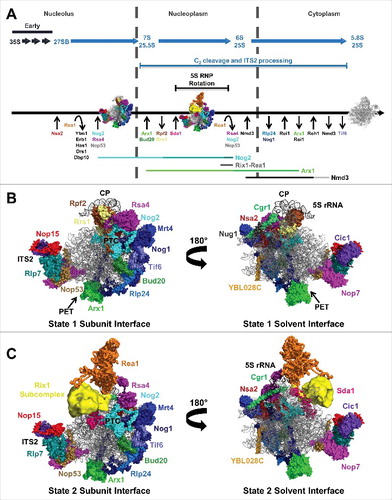
Figure 2. Yeast pre-rRNA processing and hierarchical recruitment of B-factors. (A) The pre-rRNA processing pathway in yeast. (B) Assembly of B-factors begins early in 60S subunit assembly with binding of Nip7 and Nop2 to the pre-60S subunit. The remaining B-factors assemble in 2 parallel pathways, ending with recruitment of Nog2.
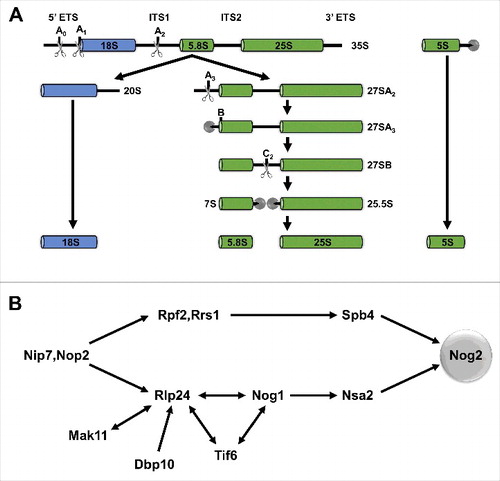
Figure 3. New insights into construction and inspection of the PTC and PET and ITS2 cleavage. (A) Nog2, Nog1, Nug1, and Nsa2 form extensive interactions with the PTC during late nuclear stages of 60S subunit assembly. Only 69 amino acids of the N-terminus of Nug1 were resolved in state 1. (B) Nog1, Rei1, and Reh1 consecutively probe the PET during 60S subunit assembly. Nog1 and Rei1 interact with Arx1 at the PET exit. (C) L25 extends from the exit of the PET to ITS2, interacting with Nop53, Nop7, Rlp7, and Nop15. Domain V of 25S rRNA stretches from the PTC to the ITS2 region. H79 of domain V interacts with Nop53 and Rlp7.