Figures & data
Figure 1. (A) Pyrrolysine is synthesized from 2 equivalents of lysine in 3 steps. (B) tRNAPyl is charged with pyrrolysine by pyrrolysyl-tRNA synthetase. (C) Pyrrolysine is incorporated into the nascent peptide in response to amber (UAG) codons during translation.
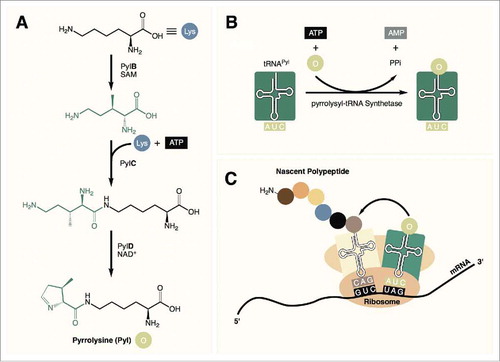
Figure 2. The secondary structure of tRNAPyl from Candidatus Methanomethylophilus alvus, Methanosarcina barkeri Fusaro, and Desulfitobacterium hafniense. Identity elements in the tRNAPyl from M. barkeri and D. hafniense are shown in orange. Bases in direct contact with the enzyme in the crystal structure of PylSc from D. hafniense are bold. In the structure from Ca. M. alvus, circles represent positions that are occupied by a base in the tRNAPyl from other Methanomassiliicoccales while underlined bases represent those that are missing.
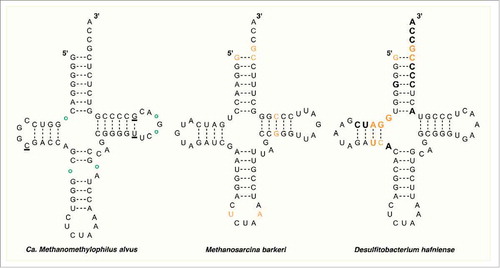
Figure 3. (A) The crystal structure of PylSc in complex with tRNAPyl from Desulfitobacterium hafniense (PDB: 2ZNI). (B) The crystal structure of tRNAPyl from D. hafniense. The secondary structure features of tRNAPyl are colored: green, acceptor stem; blue, D arm; brown, anticodon stem; purple, anticodon; yellow, variable loop; orange, T arm.
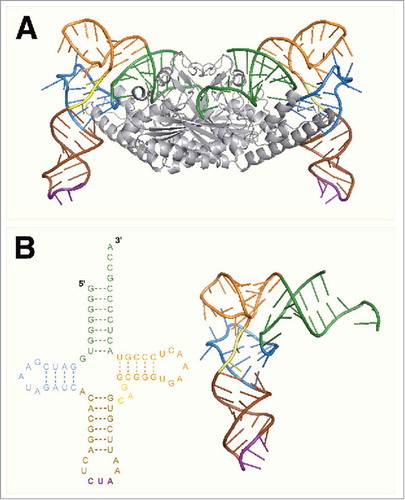
Figure 4. The crystal structure of Dh-tRNAPyl bound to Dh-PylSc. The 2 protomers in the Dh-PylSc dimer are colored gray and black. Residues that interact with Dh-tRNAPyl in the crystal structure are shown in cyan and salmon, respectively. The colors corresponding to secondary structure features of Dh-tRNAPyl are the same as in . For clarity, only one Dh-tRNAPyl is shown bound to the dimer. (PDB: 2ZNI).
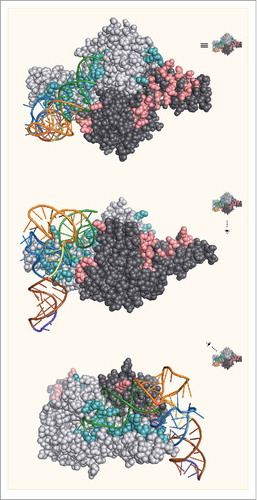
Table 1. The interactions between bases in Dh-tRNAPyl and amino acid residues in Dh-PylSc. Unless noted, interactions involve main chain hydrogen bonds.
Figure 5. (A) Mm-PylRS and Mj-TyrRS specifically charge their respective tRNAs with an ncAA. (B) Mm-tRNAPyl UUA incorporates an ncAA at ochre (UAA) codons while Mj-tRNATyr CUA incorporates ncAAs in response to amber (UAG) codons. (C) Dual labeling of glutamine binding protein containing two reactive ncAAs.
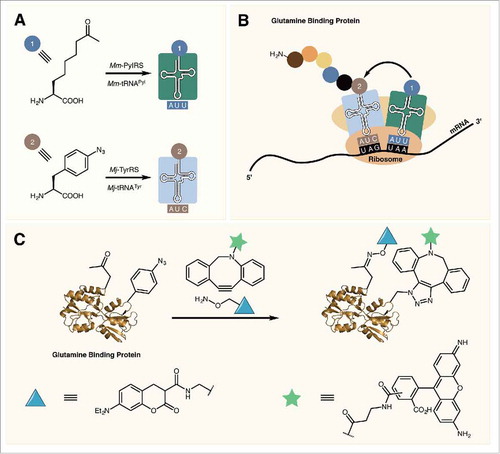
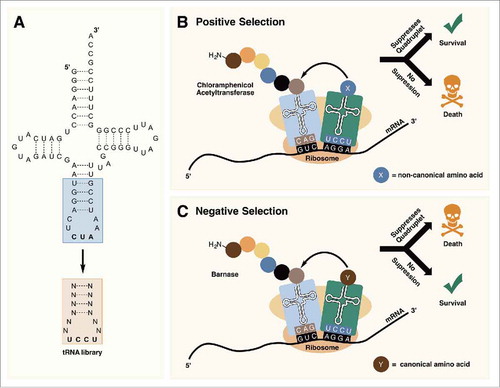
Figure 6. (A) Randomization of the anticodon stem and loop allows for the optimization of tRNAPyl for decoding quadruplet codons. (B) In positive selection, tRNAPyl mutants that are able to suppress quadruplet codons are selected for. (C) Negative selection in the absence of an ncAA removes tRNAPyl mutants that are aminoacylated with canonical amino acids by endogenous aminoacyl-tRNA synthetases.