Figures & data
Figure 1. Proposed scenarios for σ-dependent regulation of transcription elongation. (A) σ can remain bound to the TEC and induce pausing, dissociate and rebind RNAP during RNA elongation. (B) σ retention in the TEC may prevent binding of competing transcription factors, such as RfaH and NusG, which regulate transcriptional pausing and termination. (C) σ-dependent pausing serves to recruit transcription factors, such as λQ, to the TEC. (D) Promoter-proximal pausing and TEC arrest may inhibit the next round of transcription initiation and suppress recognition of cryptic promoters. (E) Promoter-proximal pausing may prevent binding of transcription repressors and facilitate continuous transcription initiation.
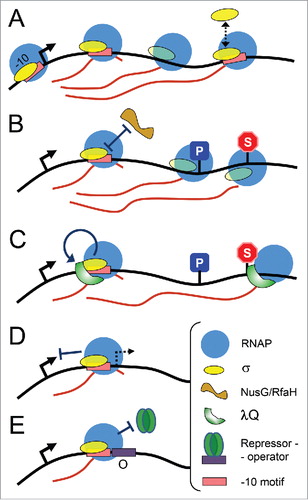
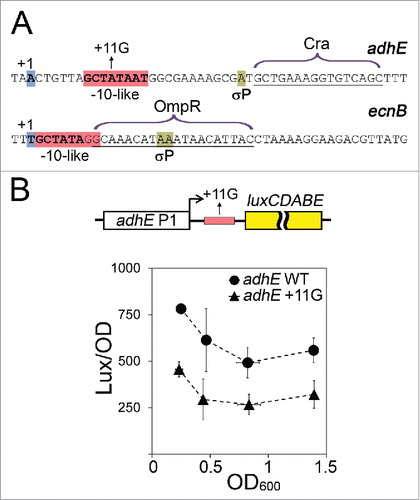
Figure 2. Possible effects of σ38-dependent pausing on repressor binding. (A) Sequences of the initially transcribed regions of adhE and ecnB promoters. The transcription start points are blue; the -10-like pause-inducing motifs are pink; the pause sites are light green; the Cra and OmpR binding sites are indicated. (B) In vivo analysis of the adhE P1 activity. E. coli cells were transformed with plasmids containing either wild-type or +11G adhE P1 variants (positions from -164 to +56 relative to the start of transcription), cloned upstream of the luxCDABE operon from Photorhabdus luminescens (shown on the top). The cultures were grown in LB medium containing 0.1 M MOPS, pH 7.4, 0.2% glucose and kanamycin in anaerobic conditions for 3, 3.5, 4 and 7 hours prior to measurements of OD600 and luminescence values. For each time interval, the luminescence was normalized to the OD values; the experiments were independently repeated three times and the OD and normalized luminescence values were averaged.