Figures & data
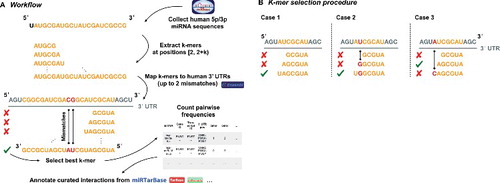
Figure 1. Schematic diagram of the reporter gene vector of TCRA. The location of the predicted binding sites and the non-canonical 5-mer binding sites of miR-34a-5p in the TCRA 3′ UTR are shown. Additionally, the diagram displays the sequences of the predicted binding sites and the non-canonical 5-mer binding sites of miR-34a-5p as well as the mutated binding sites (underlined letters).
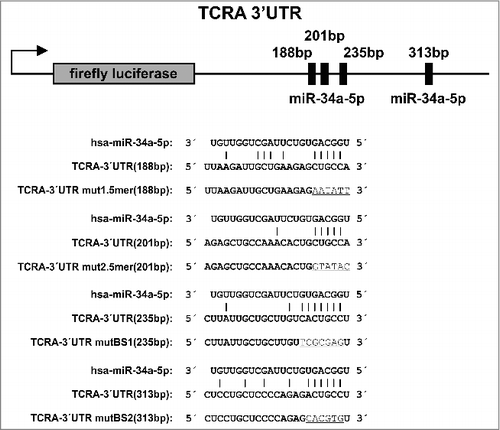
Figure 2. Number of interactions using different seed lengths and validation groups. Shown is the mean ± standard deviation of the matching miRNA-target transcript counts for each of the different k-mers (k from 5 to 8) as seed. For each k-mer length, the observed interactions are grouped, from left to right, into interactions for which no validation was known, “weak targets” identified by e.g. microarray analysis, and “strong targets” identified by e.g. luciferase reporter assays according to miRTarBase v7, TarBase v6, miRecords v4, and a large CLASH dataset [Citation52-Citation55]. The significant p-value (p = 1.4 × 10−69) between the mean of the observations in the blue (no validation) and yellow (strong evidence) group for 5-mers supports our conclusions that these small non-canonical binding sites might have a biological function in human.
![Figure 2. Number of interactions using different seed lengths and validation groups. Shown is the mean ± standard deviation of the matching miRNA-target transcript counts for each of the different k-mers (k from 5 to 8) as seed. For each k-mer length, the observed interactions are grouped, from left to right, into interactions for which no validation was known, “weak targets” identified by e.g. microarray analysis, and “strong targets” identified by e.g. luciferase reporter assays according to miRTarBase v7, TarBase v6, miRecords v4, and a large CLASH dataset [Citation52-Citation55]. The significant p-value (p = 1.4 × 10−69) between the mean of the observations in the blue (no validation) and yellow (strong evidence) group for 5-mers supports our conclusions that these small non-canonical binding sites might have a biological function in human.](/cms/asset/7ebe12fa-3e70-4f2f-9c85-98973759da6b/krnb_a_1462652_f0002_oc.jpg)
Figure 3. Dual luciferase reporter gene assay of TCRA 3′ UTR. HEK 293T cells were transfected with the indicated combinations of empty plasmids, reporter gene constructs and miRNA-34a expression vectors. 48h after transfection the cells were lysed and the luciferase activity was measured. The luciferase activity of the control vector experiment was set to 100%. The results represent the mean of four independent experiments carried out in duplicates. Three asterisks correspond to p < 0.001, two asterisks to 0.001 < p < 0.01 and NS. to not significant. Data are represented as mean±SEM. The schematic illustration on the right side shows the location of the miR-34a binding sites for each reporter construct respectively. Green boxes correspond to wild type (wt) binding sites whereas red boxes indicate mutated binding sites. The labels on top of this illustration indicate which column corresponds to which functional binding sites of miR-34a.
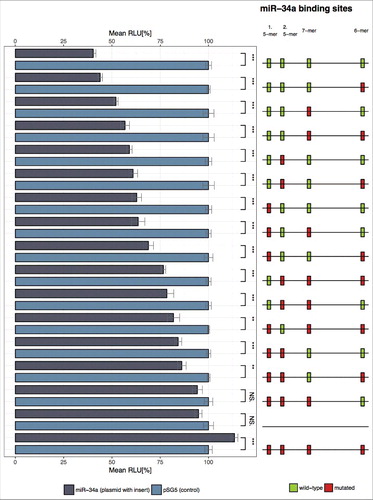
Figure 4A. Illustration on the underlying algorithm to detect miRNA k-mer enrichments in possible target sequences. Human mature miRNA 5p/3p sequences were collected from miRBase v21, while 3′ UTRs for human were acquired from Ensembl Release 91 [Citation51]. After extracting and mapping the k-mers with up to 2 mismatches for each possible binding site, the best k-mer was selected. Subsequently, for each miRNA and gene pair for which at least one valid k-mer existed we counted the number of 5-, 6-, 7-, and 8-mers. Finally, all pairs were annotated with their experimental status using validated interactions from miRTarbase v7, TarBase v6, miRecords v4, and a custom CLASH dataset [Citation52-Citation55].
![Figure 4A. Illustration on the underlying algorithm to detect miRNA k-mer enrichments in possible target sequences. Human mature miRNA 5p/3p sequences were collected from miRBase v21, while 3′ UTRs for human were acquired from Ensembl Release 91 [Citation51]. After extracting and mapping the k-mers with up to 2 mismatches for each possible binding site, the best k-mer was selected. Subsequently, for each miRNA and gene pair for which at least one valid k-mer existed we counted the number of 5-, 6-, 7-, and 8-mers. Finally, all pairs were annotated with their experimental status using validated interactions from miRTarbase v7, TarBase v6, miRecords v4, and a custom CLASH dataset [Citation52-Citation55].](/cms/asset/a3e01ca1-76d3-4d98-9b22-48fb3ec8d0d6/krnb_a_1462652_f0004a_oc.jpg)
Figure 4B. Rules for selecting the best k-mer for each possible binding site between a miRNA and a target sequence based on k-mer alignments are depicted. In case 1 the longest k-mer, in this case a 7-mer, forms the best match since no mismatches occurred and no better, i.e. longer, alignment was found. For case 2 the shown 7-mer is still the best k-mer match since a mismatch (shown in red) occurred inside the 7-mer but not at the leftmost end. In case 3 the 6-mer is selected as best matching as the 7-mer has a non-valid mismatch according to the table of allowed mismatches and the mismatch occurred at the leftmost position of the 7-mer. All other occurring alignment cases can be lead back to any of the three cases and treated analogously.