Figures & data
Figure 1. Nucleotide sequences of HSURs 1, 2, and 5 and miR-142-3p, miR-16, and miR-27a. Binding sites for miR-142-3p (highlighted in blue), miR-16 (highlighted in green), and miR-27a (highlighted in purple) are shown. Seed regions of miRNAs are highlighted in yellow. AU-rich element (ARE)-like sequences are shown in red boxes, whereas the Sm-binding site is shown in a black box.
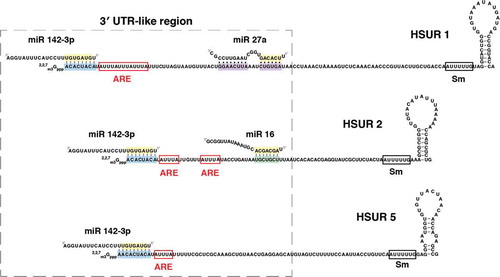
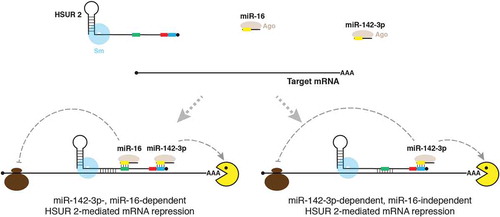
Figure 2. Model of HSUR 2 function. HSUR 2 base-pairs with a target mRNA and miRNAs miR-142-3p and miR-16 to recruit Ago proteins to the target mRNA, resulting in repression of the target mRNA. HSUR 2 could use sequences that do not involve the binding sites for miR-142-3p (blue box) and miR-16 (green box) to base-pair with the target mRNA, which would result in miR-142-3p- and miR-16-dependent mRNA repression. Alternatively, HSUR 2 could employ sequences that involve the miR-16 binding site to base-pair with the target mRNA, which would result in miR-142-3p- dependent, miR-16-independent, mRNA repression. Red box represents ARE-like sequence.