Figures & data
Figure 1. Cytoplasmic localization of RNA exosome in P.falciparum parasites.
(a) Schematic representation of constructs for generation of epitope-tagging PfRrp4 (left) and PfDis3 (right) transfectant lines by the CRISPR-Cas9 system. For both strains, the plasmids carrying a single guide RNA (sgRNA) and the Cas9 endonuclease were co-transfected. bsd, blasticidin S deaminase. hdhfr, human dihydrofolate reductase. HR, homology region.(b) Western blot analysis of Ty1-HA-PfRrp4 and PfDis3-HA-Ty1 lines with nuclear and cytoplasmic extracts, respectively. Aldolase and Histone 3 were used as loading control. CE, cytoplasmic extract. NE, nuclear extract. R, ring. T, trophozoite. S, schizont.(c) Immunofluorescence assay (IFA) of Ty1-HA-PfRrp4 and PfDis3-HA-Ty1 lines in Ring, Trophozoite, Schizont, respectively. The nuclei were stained by DAPI. The bar represents 1 μm.
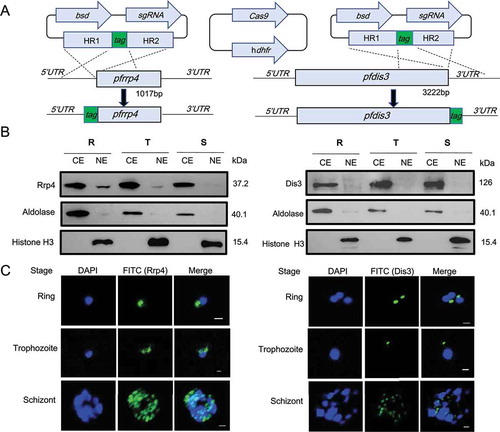
Figure 2. Composition identification of P.falciparum RNA exosome subunits.
(a) Schematic representation of exosome isolation by Co-IP with antibody against HA fused to PfRrp4 and PfDis3 respectively.(b) Comparative summary of the RNA exosome composition in Archaea, S.cerevisiae, human, and P.falciparum identified by Co-IP and LC-MS/MS in this study.(c) Putative model of RNA exosome complex of P.falciparum parasites compared with that of Archaea, S.cerevisiae, and human.
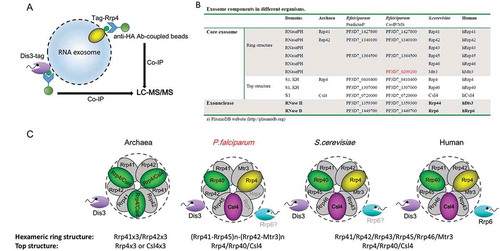
Figure 3. Generation of DiCre recombinase-mediated conditional PfDis3 KO line.
(a) Schematic representation of the PfDis3-loxPint conditional KO strategy. Co-transfection of the plasmid pL7-dis3-loxPint with pUF1-Cas9 leads to integration of the loxPint containing cassette in the endogenous locus. Activation of DiCre recombinase by rapamycin induces the excision of the loxP flanked CDS region that contains the exonuclease domain (asterisk) and by frameshift mutation to a premature termination codon (PTC, red diamond). The sgRNA target sequence is marked with SEED.(b) PCR verification of the conditional KO using oligonucleotides in homology box1 and the 3ʹCDS (p1-p2). Bands of the expected size were observed (wildtype-1020bp, undeleted Pfdis3-loxPint 1250bp, deleted Pfdis3-loxPint 610bp; gDNA harvested day 2 after induction).(c) Repeated induction and outgrowth of KO cultures. On day 2 after the first induction, rapamycin treatment was repeated and gDNA from the following days analyzed by PCR (p1-p3, expected size undeleted: 855bp, deleted: 217bp).
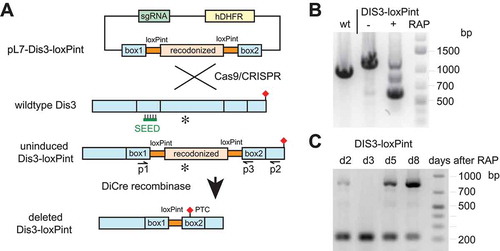
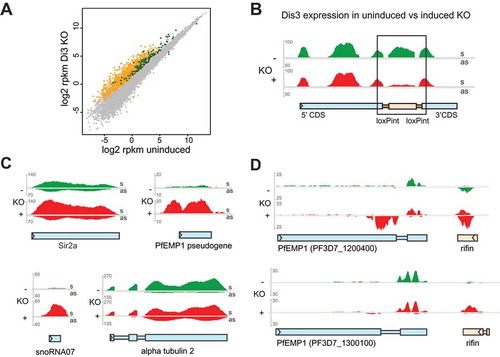
Figure 4. Differential gene expression and accumulation of virulence gene-associated cryptic unstable transcripts (CUTs) in Dis3-loxPint KO line.
(a) Scatter plot of differential gene expression in the Pfdis3-loxPint KO. Log2 RPKM values of the uninduced Pfdis3-KO are plotted against the log2 RPKM values of the induced KO (combined results from two clones in replicate). The yellow and dark green dots mark differentially expressed (≥ 1.75 fold) antisense and sense transcripts, respectively. The grey dots mark genes that are not significantly changed (combined sense and antisense).(b) Expression profile for the Pfdis3 transcript, with the black box highlighting the KO region (bedgraph coverage illustration of relative expression from IGV). The uninduced expression profile is marked in green and the Pfdis3-KO in red; upper part of the panel show sense (s) and the lower panel antisense (as) expression for both conditions, respectively.(c) Other representative examples of differentially expressed genes: increased sense transcript was observed for Sir2a (PF3D7_1328800), PfEMP1 pseudogene (PF3D7_1480100), snoRNA07 (PF3D7_1109000), while for alpha tubulin 2 (PF3D7_0422300) antisense RNA is increased.(d) Representative expression profiles of two silent var genes and their neighboring rif (PF3D7_1200500 and PF3D7_1300200). As these genes are coded on different strands, sense direction for the var gene and antisense for the rif are in the top part of the panel; antisense for the var gene and sense for the rif in the lower part.