Figures & data
Figure 1. Effect of hnRNP siRNAs on Mcl-1 splicing. (A) RNA interference was used to reduce the levels hnRNP F, H1, K and I in MCF7 cells. Combined knockdown of hnRNP F and H1 was also achieved by either combined transfection of two siRNAs (hnRNP F(1) and H1(1) siRNAs) or by using a single siRNA against a conserved region (hnRNP F/H1 siRNA). Four controls comprising a positive transfection control (GAPDH siRNA (G)), a negative scrambled sequence control siRNA (NC), a vehicle only control (V) and untreated cells (UT) were also used. Knockdown of hnRNPs was assessed at mRNA level at 48 hour using classical PCR. Alternative splicing events in MCF7 cells (B and C) and MDA-MD-231 cells (D and E) were measured using splice specific primers by qPCR at 72 hours post-transfection (mean (n = 3) ± SEM). * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001.
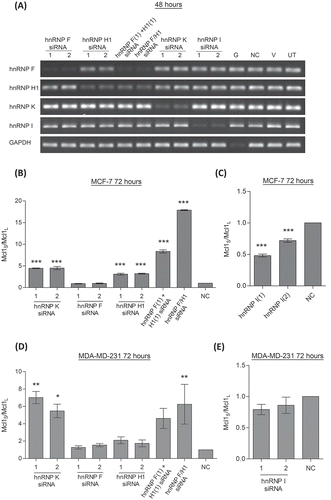
Figure 2. Binding of hnRNPs to Mcl-1 transcript. Binding of splicing factors to endogenous Mcl-1 pre-mRNA was determined by RNA immunoprecipitation using antibodies that target hnRNP H1, hnRNP K and hnRNP I. Fold enrichment of target mRNA was determined by qPCR.
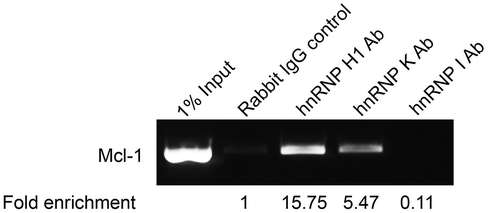
Figure 3. Identifying splicing factor binding sites using a Mcl-1 minigene. (A) Construction of the Mcl-1 minigene containing the last 115bp of exon 1, intron 1, exon 2, intron 2 and the first 157bp of exon 3. Location of minigene specific primers are indicated by arrows. Site directed mutagenesis was performed on the predicted binding sites for SRSF1 (B) and hnRNP F/H (C) within the Mcl-1 minigene. Changes to the splicing pattern were determined using the minigene specific primers and classical PCR. Bands were quantified by capillary electrophoresis (mean (n = 3) ± SEM). * P ≤ 0.05, ** P ≤ 0.01.
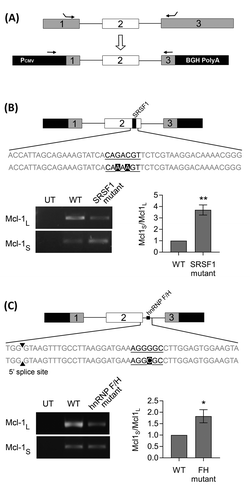
Figure 4. Double knockdowns of key Mcl-1 splicing factors. Cells were transfected with two separate siRNAs to achieve double knockdowns of key splicing factors (hnRNP K(1), hnRNPF/H1 and SRSF1 siRNA). Four controls comprising a positive transfection control (GAPDH siRNA (G)), a negative scrambled sequence control siRNA (NC), a vehicle only control (V) and untreated cells (UT) were also used. Knockdown of hnRNPs in MCF7 cells was assessed at 48 hours using classical PCR (A) and at 72 hours by western blot (B). Alternative splicing of Mcl-1 was measured in MCF7 (C) and MDA-MD-231 (D) cells using splice specific primers by qPCR at 72 hours post-transfection (mean (n = 3) ± SEM). * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001.
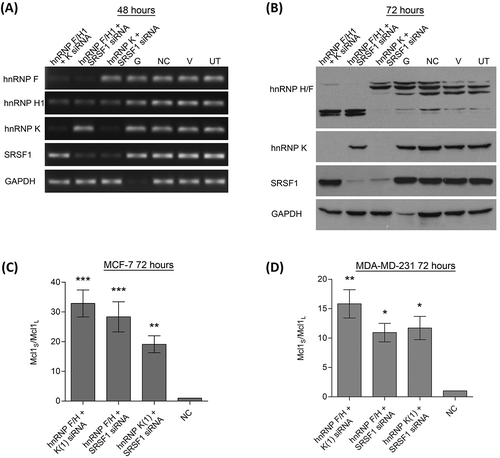
Figure 5. Induction of apoptosis by splicing factors. Cells were transfected with two separate siRNAs to achieve double knockdowns of key splicing factors (hnRNP K(1), hnRNPF/H1 and SRSF1 siRNA), a negative scrambled sequence control siRNA (NC) was used as the control. Cell viability of MCF7 (A) and MDA-MD-231 (B) cells was assessed by cell counts and trypan blue exclusion (mean (n = 3) ± SEM). (C) Caspase 9 activity was determined in the MDA-MD-231 cells 72 hours after transfection with siRNAs. Enzymatic activity of caspase 9 is expressed as fold induction compared to the control (negative control siRNA) and given as mean (n = 3) ± SEM. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001.
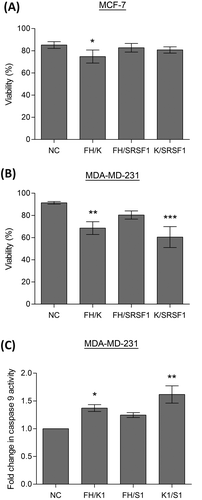
Table 1. siRNA sequences.
Table 2. PCR primers.
