Figures & data
Figure 1. The transcriptional levels of NDRG1 were inhibited by overexpressing NDRG1-OT1. MCF-7 cells were transfected with NDRG1-OT1 (2 μg) for 24 h, and NDRG1 (2 μg) for another 24 h. The expression levels of NDRG1-OT1 and NDRG1 were measured in MCF-7 cells by qRT-PCR. The relative expression level was normalized to empty control. Internal control: 18s. Data were repeated at least 3 times, and the results shown are the means ± SDs. *: P < 0.05 versus empty control.
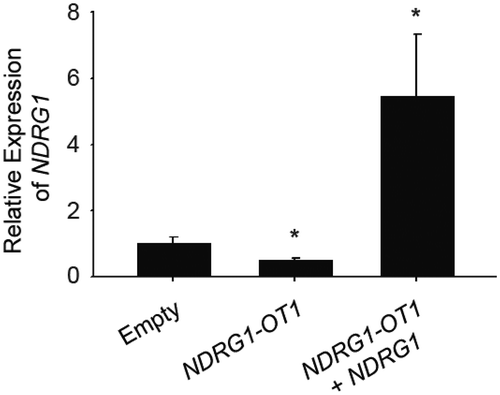
Figure 2. Different fragments of NDRG1-OT1 have different effects on NDRG1. (a) The predicted 2D structure of NDRG1-OT1 by NONCODE (http://www.noncode.org/show_rna.php?id=NONHSAT129220&version=2&utd=1#) and 3D structure of each fragment by SimRNA [Citation34]. nt: nucleotides. (b) NDRG1 promoter activity measured by luciferase reporter assays. HEK293T cells treated with CoCl2 (300 μM) were co-transfected with the Firefly luciferase, Renilla luciferase, and various fragments of NDRG1-OT1 for 24 h. Firefly luciferase activities were first adjusted by Renilla luciferase activities, and then normalized to empty control. AS: antisense. (c) Relative NDRG1 expression levels by quantitative RT-PCR. MCF-7 cells treated with CoCl2 were transfected with different fragments of NDRG1-OT1 for 24 h. The relative expression levels of NDRG1 were first normalized to internal control (18s rRNA), and then adjusted to empty control. Experiments were repeated at least 3 times, and the results shown are the means ± SDs. *: P < 0.05 as compared to empty control.
![Figure 2. Different fragments of NDRG1-OT1 have different effects on NDRG1. (a) The predicted 2D structure of NDRG1-OT1 by NONCODE (http://www.noncode.org/show_rna.php?id=NONHSAT129220&version=2&utd=1#) and 3D structure of each fragment by SimRNA [Citation34]. nt: nucleotides. (b) NDRG1 promoter activity measured by luciferase reporter assays. HEK293T cells treated with CoCl2 (300 μM) were co-transfected with the Firefly luciferase, Renilla luciferase, and various fragments of NDRG1-OT1 for 24 h. Firefly luciferase activities were first adjusted by Renilla luciferase activities, and then normalized to empty control. AS: antisense. (c) Relative NDRG1 expression levels by quantitative RT-PCR. MCF-7 cells treated with CoCl2 were transfected with different fragments of NDRG1-OT1 for 24 h. The relative expression levels of NDRG1 were first normalized to internal control (18s rRNA), and then adjusted to empty control. Experiments were repeated at least 3 times, and the results shown are the means ± SDs. *: P < 0.05 as compared to empty control.](/cms/asset/61ada82a-ae3f-4849-877c-d664f295f4da/krnb_a_1553480_f0002_oc.jpg)
Figure 3. The second quarter fragment (150–263 nt) represses NDRG1 by increasing the binding affinity of HNRNPA1 under hypoxia. (a) Gel electrophoresis of nuclear proteins after RNA pull-down assays. Nuclear proteins were extracted from MCF-7 cells growing in hypoxia for 24 h or normoxia. Biotin-labeled NDRG1-OT1 (150–263 nt) was used as the bait to pull down interacting proteins. Total nuclear proteins were visualized with silver staining. Differentially expressed proteins were isolated from the gel (area indicated by the closed bracket) and analyzed by mass spectrometry. (b) Bioinformatics prediction of docking between NDRG1-OT1 (150–263 nt) and HNRNPA1 (green). Protein 3D structure of HNRNPA1 was modeled by Phyre2 [Citation35]. The docking was predicted by HDOCK [Citation36]. nt: nucleotides. (c) Western blotting of HNRNPA1 to validate the results of pull-down assays and mass spectrometry analysis. Western blot (d) and quantification (e) of HNRNPA1 in MCF-7 cells transfected with NDRG1-OT1 plasmid. Internal control: TUBA1B. The relative expression level was normalized to empty control. (f) Luciferase activity of NDRG1 promoter activity in the absence of HNRNPA1. HEK293T cells treated with CoCl2 were co-transfected with Firefly luciferase and the second quarter fragment (150–263 nt) in the absence/presence of si-HNRNPA1 for 24 h. The results shown are the means ± SDs of at least 3 separate experiments. *: P < 0.05.
![Figure 3. The second quarter fragment (150–263 nt) represses NDRG1 by increasing the binding affinity of HNRNPA1 under hypoxia. (a) Gel electrophoresis of nuclear proteins after RNA pull-down assays. Nuclear proteins were extracted from MCF-7 cells growing in hypoxia for 24 h or normoxia. Biotin-labeled NDRG1-OT1 (150–263 nt) was used as the bait to pull down interacting proteins. Total nuclear proteins were visualized with silver staining. Differentially expressed proteins were isolated from the gel (area indicated by the closed bracket) and analyzed by mass spectrometry. (b) Bioinformatics prediction of docking between NDRG1-OT1 (150–263 nt) and HNRNPA1 (green). Protein 3D structure of HNRNPA1 was modeled by Phyre2 [Citation35]. The docking was predicted by HDOCK [Citation36]. nt: nucleotides. (c) Western blotting of HNRNPA1 to validate the results of pull-down assays and mass spectrometry analysis. Western blot (d) and quantification (e) of HNRNPA1 in MCF-7 cells transfected with NDRG1-OT1 plasmid. Internal control: TUBA1B. The relative expression level was normalized to empty control. (f) Luciferase activity of NDRG1 promoter activity in the absence of HNRNPA1. HEK293T cells treated with CoCl2 were co-transfected with Firefly luciferase and the second quarter fragment (150–263 nt) in the absence/presence of si-HNRNPA1 for 24 h. The results shown are the means ± SDs of at least 3 separate experiments. *: P < 0.05.](/cms/asset/b63ab363-a48a-47f3-b088-0811c27535a5/krnb_a_1553480_f0003_oc.jpg)
Figure 4. The third quarter fragment (264–392 nt) increases NDRG1 promoter activity by recruiting HIF-1α. (a) Luciferase expression via NDRG1 promoter activity in the absence of the HRE sequence (CGTG) at 312–315 nt. Mutation at 295–298 nt and antisense oligonucleotide (AS) are positive and negative controls, respectively. HEK293T cells treated with CoCl2 were co-transfected with the luciferase vectors, NDRG1-OT1 (264–392 nt), and NDRG1-OT1 mutants. (b) Bioinformatics prediction of docking between HIF-1α (green) and NDRG1-OT1 (264–392 nt) as well as NDRG1-OT1 mutants. (c) Relative expression levels of NDRG1-OT1 (264–392 nt) after immunoprecipitation by antibody against HIF-1α. MCF-7 cells overexpressed NDRG1-OT1 (264–392 nt) and the HIF-1α P402A/P564A mutant, which is resistant to VHL-mediated ubiquitination and degradation. Relative expression of NDRG1-OT1 (264–392 nt) was measured by qRT-PCR. The results shown are the means ± SDs of at least 3 separate experiments. *: P < 0.05.
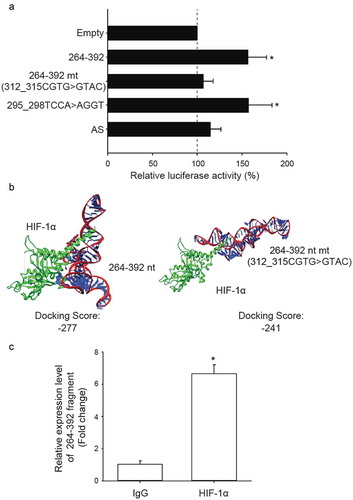
Figure 5. The fourth quarter fragment (393–508 nt) represses NDRG1 promoter activity by down-regulation of KHSRP under hypoxia. (a) Gel electrophoresis of nuclear proteins after RNA pull-down assays. The experimental conditions were the same as in . (b) Bioinformatics prediction of docking between NDRG1-OT1 (393–508 nt) and KHSRP (green). Protein 3D structure of KHSRP was modeled by Phyre2 [Citation35]. The docking was predicted by HDOCK [Citation36]. nt: nucleotides. (c) Western blotting of KHSRP to validate the results of pull-down assays and mass spectrometry analysis. (d) Luciferase expression via NDRG1 promoter activity in cells overexpressing KHSRP. HEK293T cells treated with CoCl2 were co-transfected with the Firefly luciferase and the fourth quarter fragment (393–508 nt) in the absence/presence of KHSRP for 24 h. The results shown are the means ± SDs of at least 3 separate experiments. *: P < 0.05.
![Figure 5. The fourth quarter fragment (393–508 nt) represses NDRG1 promoter activity by down-regulation of KHSRP under hypoxia. (a) Gel electrophoresis of nuclear proteins after RNA pull-down assays. The experimental conditions were the same as in Figure 2. (b) Bioinformatics prediction of docking between NDRG1-OT1 (393–508 nt) and KHSRP (green). Protein 3D structure of KHSRP was modeled by Phyre2 [Citation35]. The docking was predicted by HDOCK [Citation36]. nt: nucleotides. (c) Western blotting of KHSRP to validate the results of pull-down assays and mass spectrometry analysis. (d) Luciferase expression via NDRG1 promoter activity in cells overexpressing KHSRP. HEK293T cells treated with CoCl2 were co-transfected with the Firefly luciferase and the fourth quarter fragment (393–508 nt) in the absence/presence of KHSRP for 24 h. The results shown are the means ± SDs of at least 3 separate experiments. *: P < 0.05.](/cms/asset/dbcf0209-ca04-4eb1-a4d2-a499e6799d94/krnb_a_1553480_f0005_oc.jpg)
Figure 6. Verification of NDRG1-OT1 interaction with HNRNPA1, HIF-1α, and KHSRP using RNA pull-down assays followed by western blotting. (a) Bioinformatics prediction of docking between NDRG1-OT1 and HNRNPA1, HIF-1α, and KHSRP, respectively. The 3D structure of NDRG1-OT1 was modeled by RNAComposer [Citation52]. Color corresponds to each fragment. 2nd quarter: green; 3rd quarter: red; 4th quarter: cyan. The 3D structure of proteins (purple) was modeled by Phyre2 [Citation35]. The docking prediction was performed by HDOCK [Citation36]. (b) Western blots of HNRNPA1, HIF-1α, and KHSRP after RNA pull-down assays using full length (1–508 nt) NDRG1-OT1 as the probe. (c) Proposed model of NDRG1-OT1 interacting with various proteins to regulate the transcription of NDRG1 under hypoxia.
![Figure 6. Verification of NDRG1-OT1 interaction with HNRNPA1, HIF-1α, and KHSRP using RNA pull-down assays followed by western blotting. (a) Bioinformatics prediction of docking between NDRG1-OT1 and HNRNPA1, HIF-1α, and KHSRP, respectively. The 3D structure of NDRG1-OT1 was modeled by RNAComposer [Citation52]. Color corresponds to each fragment. 2nd quarter: green; 3rd quarter: red; 4th quarter: cyan. The 3D structure of proteins (purple) was modeled by Phyre2 [Citation35]. The docking prediction was performed by HDOCK [Citation36]. (b) Western blots of HNRNPA1, HIF-1α, and KHSRP after RNA pull-down assays using full length (1–508 nt) NDRG1-OT1 as the probe. (c) Proposed model of NDRG1-OT1 interacting with various proteins to regulate the transcription of NDRG1 under hypoxia.](/cms/asset/51be83c2-db4f-4436-8489-b77a7f2eb409/krnb_a_1553480_f0006_oc.jpg)
Table 1. The primers for quantitative RT-PCR.
