Figures & data
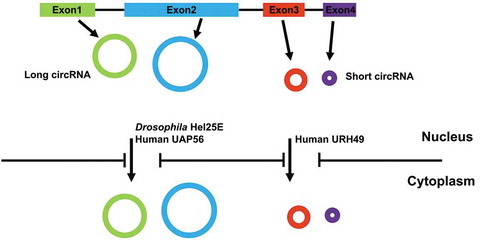
Figure 1. Nuclear export of circRNAs utilizes two separate pathways based on length. Most circRNAs, generated from exon(s), are exported to the cytoplasm. The factors required for export is determined by the size of the circRNA. In Drosophila cells, only long circRNAs require DExH/D box RNA helicase Hel25E. In human cells, one of the two human homologs of Hel25E (UAP56) similarly regulates nuclear export of long circRNAs. In contrast, URH49, the other human homolog of Hel25E, is required for nuclear export of short circRNAs.