Figures & data
Figure 1. Screening miRNAs based on porcine piPSC RNA-seq database. A. Heatmap of pluripotency associated genes in porcine iPSC lines and PEFs. B. The upregulated and downregulated genes in all three porcine iPSC lines (up panel) and the mRNA expression of upregulated pluripotent genes in PEFs and piPSC-dox cells (down panel). C. Conjoint analysis of miRNAs that potentially bind to 3ʹUTR of OTX2 by the three online programs. D. Putative binding sites in 3ʹUTR of OTX2 and the seed sequences (in red) of miRNAs. Data indicate mean ± SD. ** P < 0.01, n = 3.
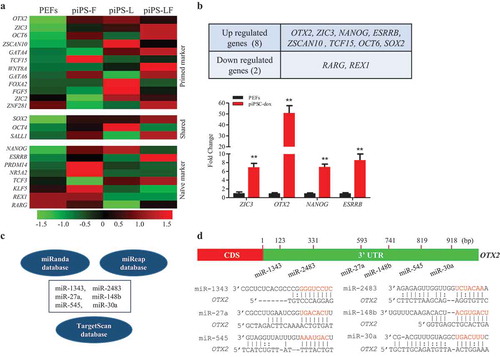
Figure 2. Validation of interactions between miRNAs and target OTX2. A. Luciferase assays of OTX2 in HEK-293T cells treated by each individual miRNA (left panel) and miRNA mimic (right panel). B. Luciferase assays of OTX2 in HEK-293T cells treated by cotransfection of miR-1343 and miR-545 (left panel) and miRNA mimics (right panel). C-D. Luciferase assays of OTX2 in HEK-293T cells with the treatments of miR-1343 mimic (C) and miR-545 mimic (D) together with their mutant (MUT) and wild type (WT) constructs. E-F. The mRNA and protein expression of OTX2 in piPSC-dox cells (E) and PS23 cells (F) treated by miRNAs. Ctrl, cells were transfected with the pCD513B-1. β-Actin was as internal control. NC-M, the negative control mimic. Data indicate mean ± SD. ** P < 0.01, n = 3.
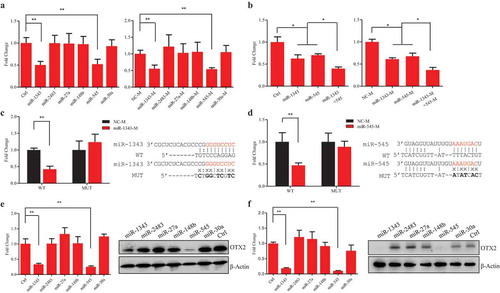
Figure 3. MiR-1343 regulates porcine iPSC pluripotency via OTX2 knockdown. The piPSC-dox and PS23 cells were treated with miRNAs or miRNA plus OTX2 vector for 72 h, and then cells were used for the following experiments. A-B. Alkaline phosphatase (AP) staining of piPSC-dox cells (A) and PS23 cells (B). Scale bar, 200 μm. C-D. The protein expression of OTX2 in piPSC-dox (C) and PS23 (D) cells. E-F. The mRNA expression of pluripotent genes in piPSC-dox (E) and PS23 (F) cells. Ctrl, cells transfected with pCD513B-1. β-Actin was as internal control. Data indicate mean ± SD. * p < 0.05, ** P < 0.01, n = 3.
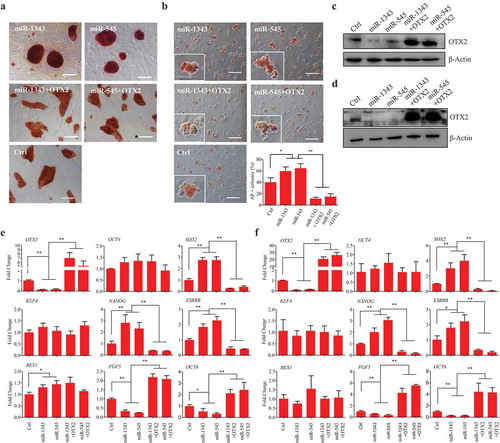
Figure 4. OTX2 represses the expression of SOX2 and ESRRB. A. Schematic diagram of the screening assay. Stem cells were transfected by miRNAs or miRNA plus OTX2 vector for 36 h. The promoter activation was then determined by luciferase assay. B. Promoter activity of pluripotent genes in P19 cells that were co-transfected with pcDNA3.1 OTX2 and reporter vector of OCT4, SOX2, and ESRRB, respectively. C-D. Dose-dependent (left panel) and time-course (right panel) assays for SOX2 (C) and ESRRB (D) promoter activity in P19 cells. Ctrl, cells were transfected by pcDNA3.1. E. Promoter activity of pluripotent genes in piPSC-dox cells. Ctrl, cells were transfected by pCD513B-1. Data indicate mean ± SD. * p < 0.05, ** P < 0.01, n = 3.
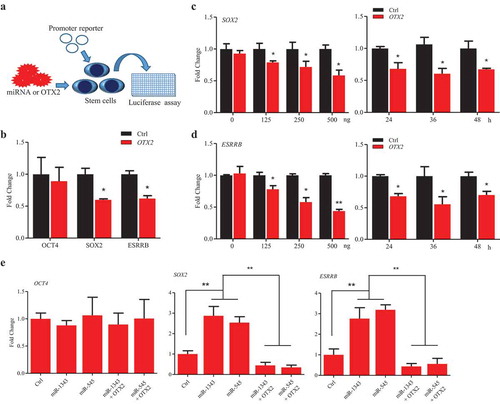
Figure 5. SOX2 represses OTX2 and stimulates miR-1343 expression. A. Luciferase assay of OTX2 promoter activity in P19 cells that were transfected by pluripotent gene constructs. B. Truncated constructs of OTX2 promoter with the predicted SOX2 binding sites. C. Luciferase assay of OTX2 promoter activity in P19 cells treated by overexpression of SOX2. D. Dose-dependent (left panel) and time-course (right panel) assays of OTX2 promoter activity in P19 cells treated by overexpression of SOX2. E-F. Pluripotent factors regulate the expression of miR-1343 and miR-545 in piPSC-dox (E) and PS23 (F) cells. Ctrl, cells were treated with pcDNA3.1 basic. Data indicate mean ± SD. * p < 0.05, ** P < 0.01 n = 3.
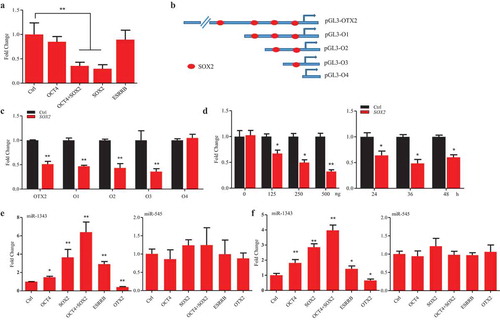
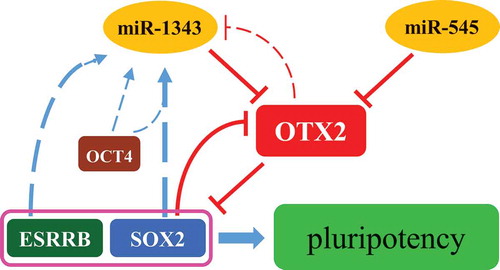
Figure 6. Regulation network of miR-1343-OTX2-SOX2 in piPSCs. The miR-1343 and miR-545 directly bind to OTX2 3ʹUTR and knock down OTX2 expression. OTX2 inhibits the expression of SOX2 and ESRRB. SOX2 is able to interacted with OTX2 promoter and represses OTX2 promoter activity. Both SOX2 and ESRRB upregulate miR-1343 expression, but OTX2 downregulates miR-1343 expression. MiR-1343-OTX2-SOX2 forms a regulatory network to maintain the pluripotency of piPSCs.