Figures & data
Figure 1. Schematic of the study flow [Citation1]. We mined miRNA candidates from next generation sequencing experiments from the literature [Citation2]. We designed a custom microarray using these candidates [11,877] and hybridized different tissues and blood to assess which candidates are more likely real miRNAs [Citation3]. From the identified high confidence miRNAs [2,267], we built a second custom microarray, which was used in the current study to measure the differential expression in patients with different diseases and controls [Citation4]. We performed statistical evaluation and added the results to our miRNA repositories and analysis pipelines [Citation5]. We selected several candidate precursors to perform validation with Northern blotting. PD = Parkinson’s disease; NSCLC = non-small cell lung cancer; SCLC = small cell lung cancer; NTLD = non-tumor lung disease.
![Figure 1. Schematic of the study flow [Citation1]. We mined miRNA candidates from next generation sequencing experiments from the literature [Citation2]. We designed a custom microarray using these candidates [11,877] and hybridized different tissues and blood to assess which candidates are more likely real miRNAs [Citation3]. From the identified high confidence miRNAs [2,267], we built a second custom microarray, which was used in the current study to measure the differential expression in patients with different diseases and controls [Citation4]. We performed statistical evaluation and added the results to our miRNA repositories and analysis pipelines [Citation5]. We selected several candidate precursors to perform validation with Northern blotting. PD = Parkinson’s disease; NSCLC = non-small cell lung cancer; SCLC = small cell lung cancer; NTLD = non-tumor lung disease.](/cms/asset/d81ea19a-b5c5-4c9a-8826-70b96d559d9e/krnb_a_1559689_f0001_c.jpg)
Figure 2. Expression density plot a: Histogram showing the distribution of the detection frequency of miRNAs annotated in miRBase (miRBase) and miRNA candidates not annotated in miRBase (new). The x-axis represents 0% to 100% of samples in intervals of 5%. The y-axis represents the relative fraction of miRNAs and miRNA candidates with the respective frequency in the 187 samples. b: Histogram showing the abundance level of miRNAs annotated in miRBase (miRBase) and miRNA candidates not annotated in miRBase (new). The x-axis represents log2 of normalized microarray expression levels. The y-axis represents the relative fraction of miRNAs and miRNA candidates with the respective expression level in the 187 samples.
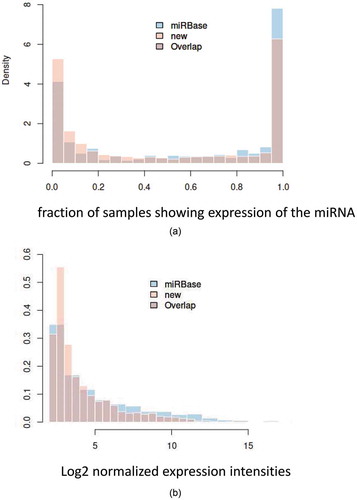
Figure 3. Volcano plots for the series of comparisons. For each comparison, the log2 fold change is shown on the x-axis and the negative decadic logarithm of the raw p-value (Wilcoxon rank sum; two-tailed) is shown on the y-axis. Nominally significant miRNAs are highlighted in red or green, depending on whether their expression levels are downregulated or upregulated, respectively, in the second comparison group compared with the first. P-values that were significant after adjustment are shown in dark green and red. The volcano plots on top shown broader comparisons, while the scatter plots at the bottom show results for specific comparisons. NTLD = non-tumor lung disease; LC = lung cancer; PD = Parkinson’s disease; LD = lung disease; SCLC = small cell lung cancer; NSCLC = non-small cell lung cancer.
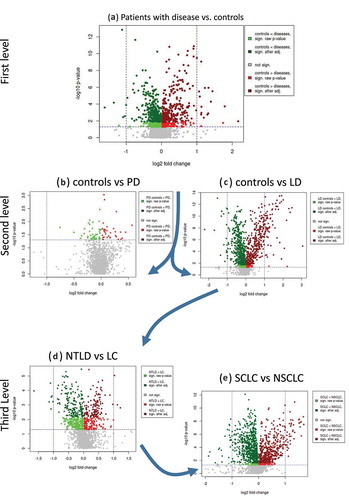
Table 1. The top 10 significant differentially expressed miRNAs/miRNA candidates in patients with disease (all diseases combined, n = 116) compared with controls (all controls combined, n = 71).
Table 2. The top 10 significant differentially expressed miRNAs/miRNA candidates in controls (n = 37) versus lung diseases (n = 82).
Figure 4. Boxplots resulting from an ANOVA. a: Boxplots for miRNA-30c-5p in four groups of patients. This miRNA was significantly downregulated in patients of all diseases compared with unaffected controls. b: Boxplots for miRNA-425-5p in four groups of patients. This miRNA showed the most significant downregulation in NSCLC patients. c: Boxplots for m-3254 in four groups of patients. This miRNA, which is not annotated in the miRBase, was most significantly upregulated in SCLC patients. L-HC = matched controls for the lung cancer samples; NTLD = non-tumor lung disease; SCLC = small cell lung cancer; NSCLC = non-small cell lung cancer.
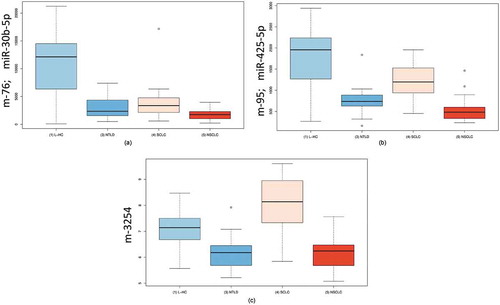
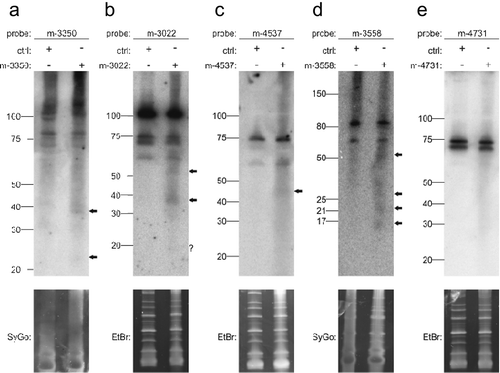
Figure 5. Northern blot detection of miRNA candidates m-3350 (a), m-3022 (b), m-4537 (c), m-3558 (d) and m-4731 (e) using radiolabeled probes in HEK293T cells transfected with the respective overexpression constructs. Ctrl indicates transfection of pSG5 vector alone. SYBR Gold (SyGo) or ethidium bromide (EtBr) was used to confirm equal loading. Markers were used to confirm RNA ladders sizes (shown on the left of each gel). Arrows indicate specific signals.