Figures & data
Figure 1. lncRNA mediated PRC2 binding. RNA immunoprecipitation studies showed the first association of RNA with PRC2. The binding of lncRNA with PRC2 was thought to be the reason behind PRC2 recruitment, and this gave rise to the idea of lncRNA specific binding of PRC2. lncRNA guided PRC2 to the target gene locus where it would identify repressive chromatin markers (H3K27me3/H2AK119ub) and cause gene repression. PRC2 was seen to deposit H3K27me3 on chromatin and cause gene repression either in cis (A) as was seen in case of Xist and RepA for XCI and in trans (B) as was seen in HOTAIR-mediated suppression of HOXD locus.
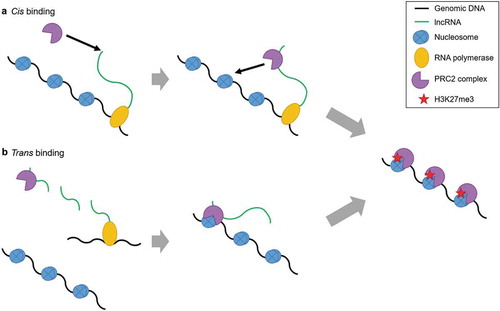
Figure 2. Model systems for RNA-mediated recruitment of PRC2. PRC2 was shown to have promiscuous binding rather than having any specificity for lncRNAs. The promiscuous nature of PRC2 was thought to be a way for PRC2 to bind with RNA and scan the genome for genes that escaped silencing (scanning), and in active genes elongating RNA acts as a decoy for PRC2 (eviction). PRC2 remained in a deactivated/poised state when bound to RNA with its HMTase activity inhibited and would deposit H3K27me3 only when it recognized repressive markers (H3K27me3/H2AK119ub). This was known as the ‘junk mail’ model (A). Due to the odds in binding seen in vitro and in vivo, it was thought that the intrinsic promiscuity of PRC2 to bind with RNA meant that it must compete with other RBPs. RNAs that were not bound to any other proteins had a higher chance to bind with PRC2. This was known was the ‘masking model’ (B). Recent iCLIP studies showed that the RNA and chromatin compete with each other to bind with PRC2, thus having an antagonistic relationship (C). In moderately active gene (C, middle) both RNA and chromatin compete, but in active genes (C, bottom) the continuous formation of RNA drives PRC2 from chromatin while in repressed genes (C, top) loss of RNA cause the chromatin to bind with PRC2.
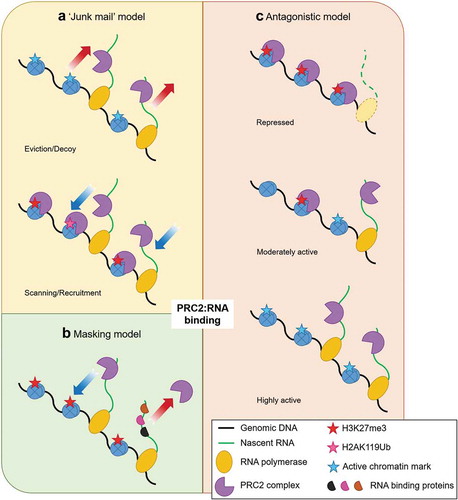
Figure 3. RBPs modulating the binding of PRC2 on genome. Several evidences have pointed out the association of RBPs with RNA and their role in regulating gene expression. RBFox2, a pre-mRNA splicing regulator, was found to be associated with nascent RNA and helped in the recruitment of PRC2 to active genes by protein-protein interaction. RBFox2 may act as a part of a complex or it may be one of the several RBPs that binds with PRC2.
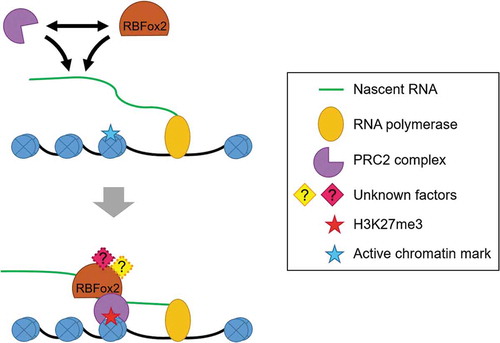
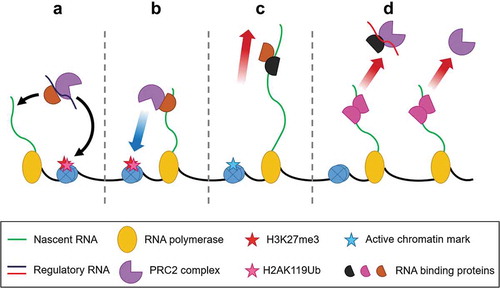
Figure 4. Speculated RNA:RBP:PRC2 interactions. The interactions between PRC2 and RNA can be postulated to be modulated by proteins that have affinity for PRC2 and RNA. In repressed genes, the proteins binding to a regulatory RNA can bring PRC2 to the genetic loci for trans repression (A) or PRC2 can bind to the proteins with nascent RNA and deposit H2K27me3 (B). In the case of active genes, even if PRC2 binds with RBPs, the elongating RNA takes PRC2 away from the chromatin and the active chromatin marks does not allow chromatin binding (C). Moreover, the proteins bound with the nascent RNA can cause a steric hindrance for PRC2 to bind or that the proteins bound to nascent RNA can inhibit PRC2 joining (D).