Figures & data
Figure 1. The Type IV Cas6 from M. australiensis (Ma Cas6-IV) cleaves the CRISPR repeat of the Type IV associated CRISPR. (a) M. australiensis contains a Type I CRISPR system and a Type IV CRISPR system, each with a distinct repeat sequence designated as Repeat I and Repeat IV. Nucleotides highlighted in blue indicate a purine-purine or pyrimidine-pyrimidine shift in sequence. Bases colored red indicate broader mutations. (b) Ma Cas6-IV-mediated processing of a pre-crRNA composed of four Repeat IV sequences interspersed with the first spacer sequence from the M. australiensis Type IV CRISPR. (c) SYBR Gold stained gel of the 30 nt long Repeat IV and the G22 2ʹ-deoxy repeat incubated with Ma Cas6-IV. The G22 2ʹ-deoxy repeat remains uncleaved in the presence of Ma Cas6-IV (d) Fit to cleavage data of a single radiolabeled repeat by Ma Cas6-IV. Data for the Repeat IV cleavage were fit as described in [Citation24]. Error bars denote standard deviation between two or more experiments. (e) Ma Cas6-IV-mediated cleavage trials of 5ʹ-32P-end labeled Repeat IV, Repeat I, and their reverse complements (Repeat IV-RC, Repeat I-RC). Data were not fit for Repeat I or the RC sequences because no cleavage was observed
![Figure 1. The Type IV Cas6 from M. australiensis (Ma Cas6-IV) cleaves the CRISPR repeat of the Type IV associated CRISPR. (a) M. australiensis contains a Type I CRISPR system and a Type IV CRISPR system, each with a distinct repeat sequence designated as Repeat I and Repeat IV. Nucleotides highlighted in blue indicate a purine-purine or pyrimidine-pyrimidine shift in sequence. Bases colored red indicate broader mutations. (b) Ma Cas6-IV-mediated processing of a pre-crRNA composed of four Repeat IV sequences interspersed with the first spacer sequence from the M. australiensis Type IV CRISPR. (c) SYBR Gold stained gel of the 30 nt long Repeat IV and the G22 2ʹ-deoxy repeat incubated with Ma Cas6-IV. The G22 2ʹ-deoxy repeat remains uncleaved in the presence of Ma Cas6-IV (d) Fit to cleavage data of a single radiolabeled repeat by Ma Cas6-IV. Data for the Repeat IV cleavage were fit as described in [Citation24]. Error bars denote standard deviation between two or more experiments. (e) Ma Cas6-IV-mediated cleavage trials of 5ʹ-32P-end labeled Repeat IV, Repeat I, and their reverse complements (Repeat IV-RC, Repeat I-RC). Data were not fit for Repeat I or the RC sequences because no cleavage was observed](/cms/asset/feba6141-4e70-483b-8f60-520afdfb0ec8/krnb_a_1634965_f0001_c.jpg)
Table 1. Data collection and refinement statistics
Figure 2. Structure of apo Ma Cas6-IV and identification of the active site residues, His44 and Tyr31. (a) Ribbon model (left) and topology diagram (right) of the Ma Cas6-IV structure. The two RRM domains, C-terminal structural motifs involved in binding the crRNA, and putative active site are indicated. (b) Alignment of apo Ma Cas6-IV to RNA-bound TthCas6 (PDB:4C8Z) with a zoomed in view of the Ma Cas6-IV active site containing His44 and Tyr31 residues. RMSD of the alignment is 1.26 Å over 95 out of 257 Cα carbons. The bottom panels show surface renditions of Ma Cas6-IV aligned with the RNA of PDB 4C8Z. Predicted positively charged surface is colored blue and negatively charged surface is colored red. (c) Cleavage assays of Repeat IV by predicted active site mutants of Ma Cas6-IV (left) with cleavage percentages fit to a pseudo-first order rate equation (right). Cleavage activity of H44A Ma Cas6-IV in the presence of 500 mM imidazole is also shown
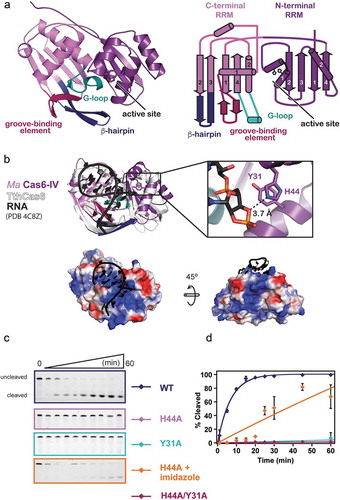
Figure 3. Ma Cas6-IV exhibits single-turnover characteristics for Repeat IV. (a) Cleavage of Repeat 4 by Ma Cas6-IV with a Cas6:Repeat IV ratio of 2:1. (b) Isotherms and electrophoresis gels of additional ratios of Cas6:Repeat are shown. The data were fit as described in [Citation24]. Error bars denote standard deviation between three experiments
![Figure 3. Ma Cas6-IV exhibits single-turnover characteristics for Repeat IV. (a) Cleavage of Repeat 4 by Ma Cas6-IV with a Cas6:Repeat IV ratio of 2:1. (b) Isotherms and electrophoresis gels of additional ratios of Cas6:Repeat are shown. The data were fit as described in [Citation24]. Error bars denote standard deviation between three experiments](/cms/asset/702578c7-f6c2-48a0-96b5-c0978ec336e8/krnb_a_1634965_f0003_c.jpg)
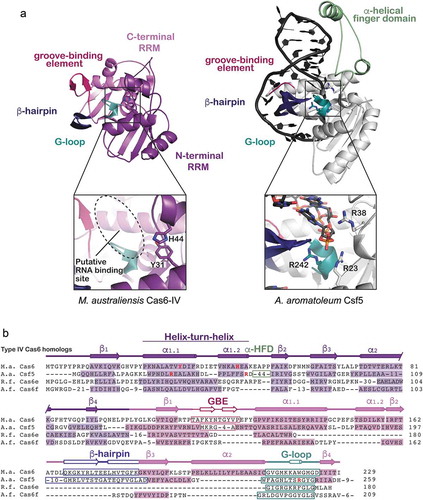
Figure 4. Structure and sequence alignments of Ma Cas6-IV with other Type IV RNA endonucleases. (a) A structural comparison of Ma Cas6-IV with the Cas6-homolog Csf5 from Aromatoleum aromaticum (PDB 6H9I). Features involved in binding crRNAs are indicated. The Csf5 protein contains a large insert called the alpha-helical finger domain (light green) that is not observed in Ma Cas6-IV. Residues predicted to activate cleavage of the crRNA are indicated in the inset below. (b) Sequence alignment of Ma Cas6-IV with other RNA endonucleases observed in Type IV systems. The N- and C-terminal RRM secondary structure elements are indicated, as well as features which bind crRNA, including the groove-binding element (GBE), beta-hairpin, and glycine-rich loop (G-loop). The alpha-helical finger domain (α-HFD) insert of Csf5 is also indicated. Active site residues of Ma Cas6-IV and Csf5 are bolded in red. Cas6e and Cas6f sequences are noticeably shorter than Ma Cas6-IV and Csf5, lacking large portions of the C-terminal RRM