Figures & data
Figure 1. Expression analysis of a pool of lncRNAs during mice NSCs differentiation
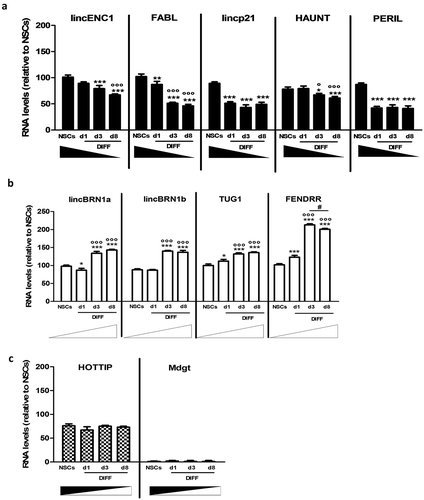
Figure 2. Silencing of lincBRN1a and lincBRN1b in NSCs
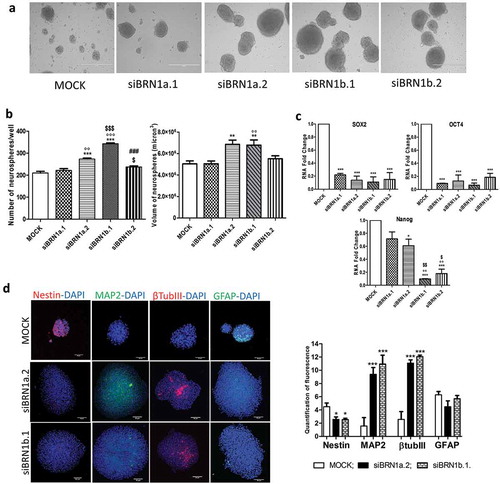
Figure 3. Silencing of lincBRN1a and lincBRN1b in differentiated NSCs
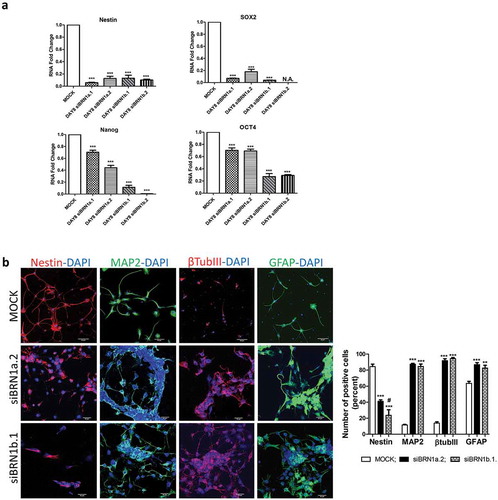
Figure 4. HuR’s expression in mouse brain stem cell niches and during mice NSCs differentiation in vitro
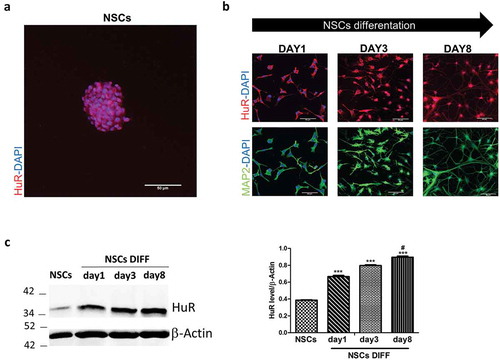
Figure 5. LncRNAs and HuR interact and this interaction influences lncRNAs’ expression
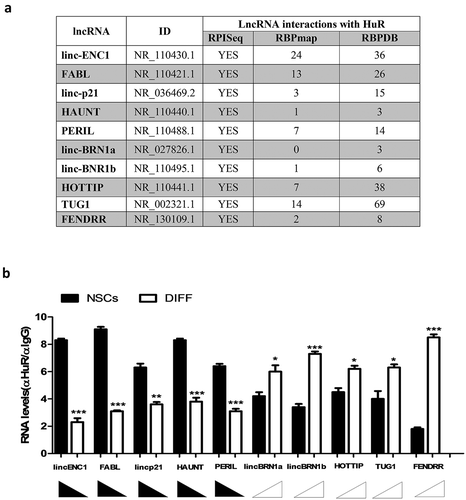
Figure 6. HuR’s inhibition and silencing in neurospheres and in differentiated NSCs
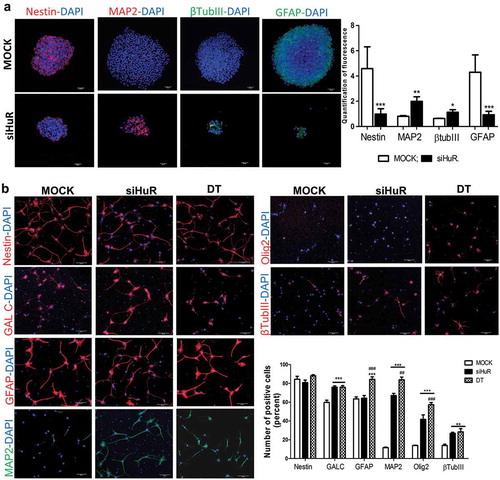
Figure 7. Investigation of the interaction between HuR and lincBRN1a/lincBRN1b
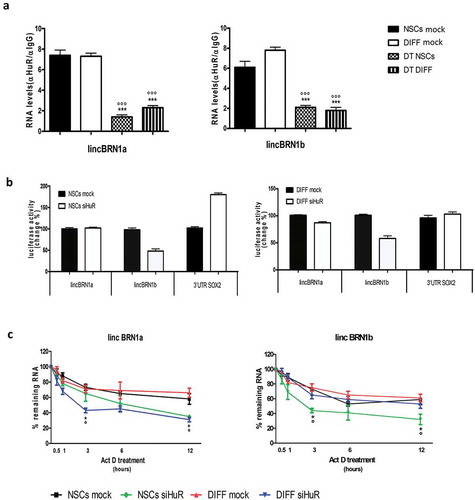
Table 1. Identification of human homologues of murine lncRNAs
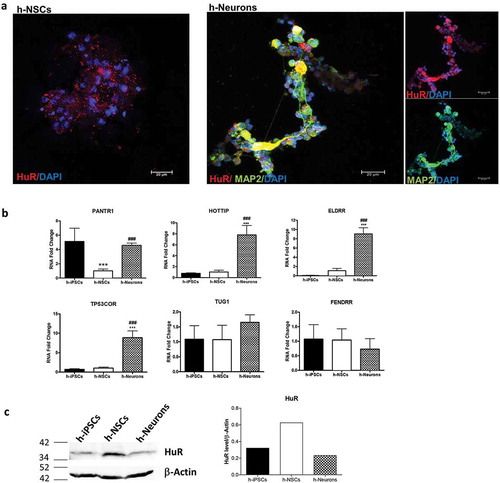
Figure 8. Expression of HuR and human homologues of lncRNAs during neuronal differentiation of h-iPSCs