Figures & data
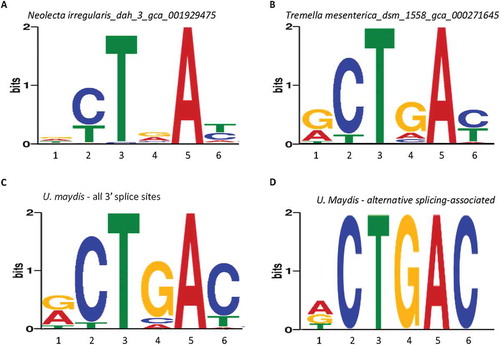
Figure 1. Representative branch point motifs identified among the Ensembl-annotated fungal species using MEME. A. Consensus of the fungal branchpoint motifs in the genomes and complementarity of the corresponding pre-mRNA motifs with U2 snRNA without or with wobble base-pairing nucleotides (Top), and three representative sequence logos of the information content (bits) of the enriched branchpoint motifs between the −30 and +2 positions of annotated 3ʹ splice sites in the genome of each species, from MEME analysis. The big black dot indicates the branchpoint nucleotide and the smaller ones the wobble pairings. B. Per cent distribution of the branch point motifs of extreme examples of G4/A4 levels in the consensus sequences of two fungal species. Percentages of G4: of all 3ʹ splice sites containing the MEME branchpoint motif in a species. The nucleotide positions are numbered according to the consensus sequence in A.
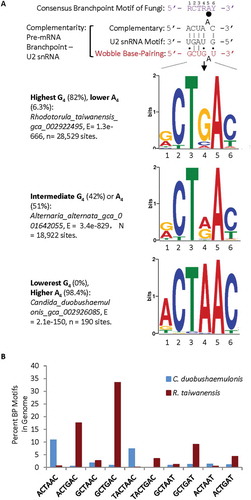
Figure 2. Distinct distribution of G4 and A4 among species/strains of different complexity. A. G4 and A4 percentages of 683 Ascomycota and 197 Basidiomycota species/strains, ranked by increasing G4%. Pezizomycotina comprises 99% of the subphyla of 475 multicellular Ascomycota species within the G4 range between 0.125 and 5.0. Note that the sudden decrease of A4 and increase of G4 separates multicellular from unicellular Ascomycota. B. Percentages of uni- or multi-cellular Ascomycota species with different thresholds of G4/A4 ratios (n = 212 unicellular, and 467 multicellular species). The ovals represent uni- or multi-cellular Ascomycota species. The Basidiomycota group also contains both uni- and multi-cellular species, but they are not clearly separated by their G4/A4 ratios. *: a group of C. neoformans strains with similar percentages of G4 or A4.
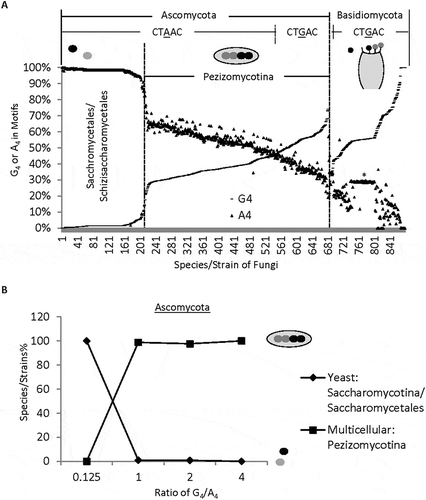
Figure 3. Relationship between the G4/A4 ratio and the total number of 3ʹ splice sites of different genomes and the G4 – A4 evolvement among different species. A. G4/A4 ratio versus the number of 3ʹ splice sites in each of 503 fungal species (395 Ascomycota, 108 Basidiomycota), in logarithmic scales. Blue markers boxed in grey-dotted line: unicellular Ascomycota species. Grey markers: C−3/T−3 (spades) or G−4/A−4 (dots) ratios of the 3ʹ splice site of 470 or 488 fungal genomes as controls for comparison. B. The G4 or A4 within the potential branchpoint motifs of a 3ʹ splice site of the conserved eIF-2B beta gene in different species/strains. The homology tree is according to the eIF-2B beta proteins (with protein IDs) aligned by ClustalW. Note that both S. Complicata strains contain duplicated eIF-2B beta genes. C. The branchpoint MEME motifs of four protist species also containing G4 and/or A4. Note that the E. invadens branchpoint A has a fixed position (−8) relative to the 3ʹ AG, and the C. paramecium consensus has extra nucleotides beyond the 6 positions focused in this study. Black dot: the branchpoint A. The nucleotide positions are numbered according to the consensus sequence in .
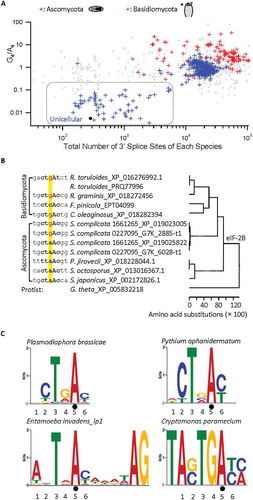
Figure 4. Distribution of the nucleotides of the 5ʹ splice site and the U2 snRNA gene entries among different fungal genomes. A. Representative distribution of the nucleotides of the 5ʹ splice site among the different fungal genomes as the branchpoint motif G4 increases or A4 decreases. Shown are A3, G3 or A4 within the MEME motif of the 5ʹ splice site. B. Bar graph showing the distribution of 1,336 unique U2 gene entries in the Rfam database with different motifs among 361 of the Ascomycota or Basidiomycota species with branchpoint G4/A4 ratios in .
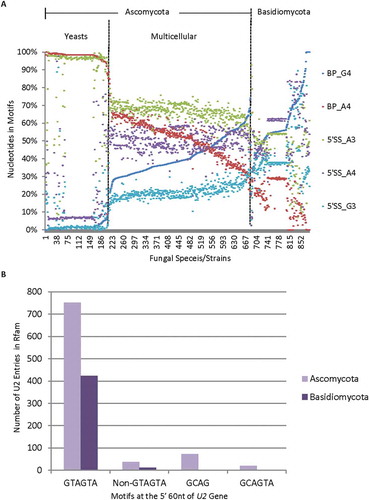
Figure 5. Relationship of the percentages of genes with at least one alternative splicing event to the G4/A4 ratios of different fungal species/phyla. Here the unicellular species of Ascomycota is S. pombe. The other (multicellular) Ascomycota species are: A. oryzae, A. flavus, F. graminearum, T. melanosporum, P. brasiliensis Pb01, C. immitis, P. brasiliensis Pb18, P. brasiliensis Pb03, A. niger, N. crassa, A. nidulans, P. anserina, P. nodorum (n = 13 species/strains). The three Basidiomycota species are: C. neoformans, S. commune, C. cinerea (n = 3). The abundance of alternative splicing in each species can be found in the references in the text. The points with error bars represent the mean (± SEM) values of each axis. The equation of the dotted trendline with the Pearson correlation coefficient is based on the mean values of the points.
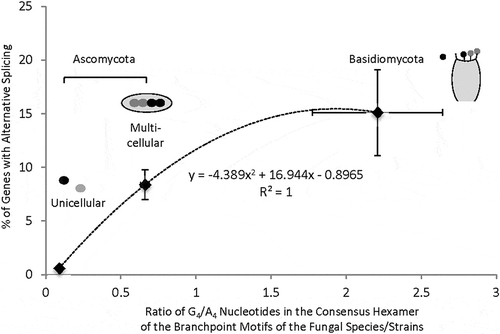
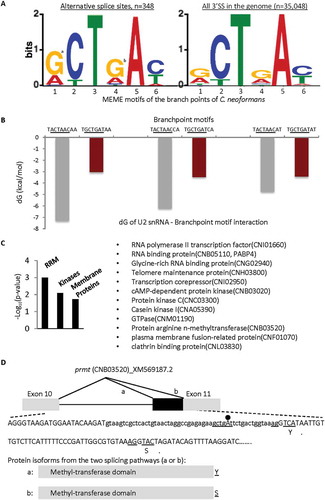
Figure 6. Enrichment of G4 and other wobble nucleotides in the MEME branchpoint motifs of the alternative 3ʹ splice sites (A), its potential effect on U2 snRNA binding (B) and functional impact (C-D), in Basidiamycota C. neoformans. The nucleotide positions are numbered according to the consensus sequence in as in previous figures. a: p= 1.3E-13, b: p= 2.6E-07, compared to the genome-wide 3ʹ splice sites, in hypergeometric test. The binding strength to the GUAGUA motif of U2 snRNA was measured by the changes in free energy dG (kcal/mol) upon transition from A4 to G4 and other nucleotides (G1 or T6) within the different branchpoint motifs of the alternative 3ʹ splice sites. The functional clusters were obtained using DAVID. Shown in D are the alternative 3ʹ splice sites of the prmt gene between exons 10 and 11 (boxes) with details of the sequence features including the branchpoint motif, 3ʹ AGs and the splice variants’ last codons (underlined, with the coded amino acids under them), as well as the variant protein domains and terminal amino acids from the two splicing pathways (a or b).