Figures & data
Figure 1. Ratio of stalled reads (RSR) metric improves resolution and specificity for detecting RTS events. (A) Showcases comparing proposed ratio of stalled reads (RSR) metric with coverage drop signal (CDS) metric used to resolve RNA G-quadruplex (rG4)-induced reverse transcriptase stalling (RTS) events. Signal peaks indicated by CDS or RSR metric were annotated with hash (#) or asterisk (*) symbols respectively. → In the 1st showcase at the BASP1 gene, the RTS event associated with the canonical rG4 was detected by both CDS and RSR. While both metrics indicated a signal peak at the 3′ end of the rG4 motif, RSR produced a sharper and narrower peak. → In the 2nd showcase at the EEF2 gene, the RTS event was detected by both CDS and RSR. This rG4 motif exhibited 5 G tracts and utilized either the 4th or 5th G tract as the 3′-most G tract. The RSR metric offered higher resolution and indicated both RTS sites corresponding to the 4th/5th G tract with two separate peaks → In the 3rd showcase at the RPL13 gene, the RTS event was detected by RSR but not CDS owing to low RTS effect strength as nucleotide positions of CDS <0.2 were removed to eliminate low-confidence data points. Compared to CDS, the RSR metric offered better resolution for closely adjacent RTS events or weak RTS events.(B) Re-analysis of previously reported RTS sites under the ‘Others’ category using the RSR metric. RTS sites under the ‘Others’ category are considered false positives and should be minimized in rG4-seq analysis. The proposed scheme rejected most of these RTS sites by criteria of insufficient read coverage (<6x), absence of associated reproducible RSR peaks, or disagreements between rG4-seq (K+) and rG4-seq (K+-PDS) experiments. Meanwhile, 26 ‘Others’ RTS sites met all detection criteria, where some were genuine rG4s incorrectly classified as false positives.
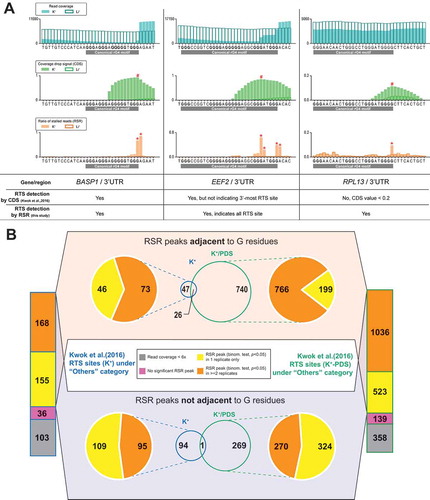
Table 1. False-positive RTS site detection is associated with low read coverage.
Figure 2. Minimum ΔRSR scheme out-performs binomial test in RTS event detection at lower read coverage. (A) The effects of low read coverage on ratio of stalled reads (RSR) signal simulated by exponential read subsampling, demonstrated by a 50 nt non-rG4-harbouring region on the RPS18 gene. At the original read coverage, an equivalent RSR signal was recorded by rG4-seq at K+ (rG4-stabilizing) and Li+ (rG4-non-stabilizing) conditions without statistically significant differences (binomial test), suggesting that the underlying RT stalling probabilities did not differ between the two conditions. However, the shape of the original RSR signal was distorted and the similarity of RSR signal between the two conditions was lost at reduced read coverage (~300x and ~30x) when under-sampling error affected the RSR measurements. (B) Comparison between the binomial test and proposed minimum ΔRSR metric scheme. A marginal case at low read coverage (~30x) extracted from a non-rG4-harbouring region in rG4-seq dataset is shown. Minimum ΔRSR metric scheme additionally addresses sampling error of RSR[K+] measurement with binomial proportional confidence interval. (C) Re-analysis of previously reported RTS sites under the ‘Others’ category using minimum ΔRSR scheme. RTS sites under the ‘Others’ category are considered false positives and should be minimized in rG4-seq analysis. The minimum ΔRSR scheme out-performed the binomial test in rejecting more ‘Others’ RTS sites and reported less reproducible RSR peaks non-adjacent to G residues. Meanwhile, 21 ‘Others’ RTS sites met all detection criteria and were adjacent to G residues.
![Figure 2. Minimum ΔRSR scheme out-performs binomial test in RTS event detection at lower read coverage. (A) The effects of low read coverage on ratio of stalled reads (RSR) signal simulated by exponential read subsampling, demonstrated by a 50 nt non-rG4-harbouring region on the RPS18 gene. At the original read coverage, an equivalent RSR signal was recorded by rG4-seq at K+ (rG4-stabilizing) and Li+ (rG4-non-stabilizing) conditions without statistically significant differences (binomial test), suggesting that the underlying RT stalling probabilities did not differ between the two conditions. However, the shape of the original RSR signal was distorted and the similarity of RSR signal between the two conditions was lost at reduced read coverage (~300x and ~30x) when under-sampling error affected the RSR measurements. (B) Comparison between the binomial test and proposed minimum ΔRSR metric scheme. A marginal case at low read coverage (~30x) extracted from a non-rG4-harbouring region in rG4-seq dataset is shown. Minimum ΔRSR metric scheme additionally addresses sampling error of RSR[K+] measurement with binomial proportional confidence interval. (C) Re-analysis of previously reported RTS sites under the ‘Others’ category using minimum ΔRSR scheme. RTS sites under the ‘Others’ category are considered false positives and should be minimized in rG4-seq analysis. The minimum ΔRSR scheme out-performed the binomial test in rejecting more ‘Others’ RTS sites and reported less reproducible RSR peaks non-adjacent to G residues. Meanwhile, 21 ‘Others’ RTS sites met all detection criteria and were adjacent to G residues.](/cms/asset/2c3dc65b-4b35-4f28-a51a-7cf31f4730d8/krnb_a_1740470_f0002_c.jpg)
Figure 3. False-positive RTS detections originate from RNA fragmentation-associated background noise. (A) Transcriptome-wide significant RSR peaks (minimum ΔRSR >0) detected from replicate 1 of HeLa rG4-seq dataset (pairwise comparison between K+/Li+ condition). Minimum ΔRSR values (indicating RTS effect strength) of RSR peaks were plotted against read coverage (logarithmic scale). Each datapoint corresponds to 1 RSR peak at 1 single-nucleotide genomic locus. RSR peaks coinciding with RTS sites in canonical/G3L1-7 reported by Kwok et al. [Citation9] are highlighted. (B) Transcriptome-wide significant RSR peaks (minimum ΔRSR >0) detected from a pairwise comparison between replicates 1 and 2 of the Li+ HeLa rG4-seq dataset. Comparison between replicates of identical conditions implies that all detected RSR peaks originated from experimental variations. (C) Summary of different reverse transcription (RT) termination events expected in rG4-seq that would generate RSR signals, and whether these events were controlled by rG4-seq (Li+) experiments. (D) Illustration of noise introduced to rG4-seq sequencing data by the RNA fragmentation process. Each rG4-seq library receives an unidentical assortment of RNA fragments, which causes discrepancies in the RT termination events at 5′ of the RNA fragments between libraries.
![Figure 3. False-positive RTS detections originate from RNA fragmentation-associated background noise. (A) Transcriptome-wide significant RSR peaks (minimum ΔRSR >0) detected from replicate 1 of HeLa rG4-seq dataset (pairwise comparison between K+/Li+ condition). Minimum ΔRSR values (indicating RTS effect strength) of RSR peaks were plotted against read coverage (logarithmic scale). Each datapoint corresponds to 1 RSR peak at 1 single-nucleotide genomic locus. RSR peaks coinciding with RTS sites in canonical/G3L1-7 reported by Kwok et al. [Citation9] are highlighted. (B) Transcriptome-wide significant RSR peaks (minimum ΔRSR >0) detected from a pairwise comparison between replicates 1 and 2 of the Li+ HeLa rG4-seq dataset. Comparison between replicates of identical conditions implies that all detected RSR peaks originated from experimental variations. (C) Summary of different reverse transcription (RT) termination events expected in rG4-seq that would generate RSR signals, and whether these events were controlled by rG4-seq (Li+) experiments. (D) Illustration of noise introduced to rG4-seq sequencing data by the RNA fragmentation process. Each rG4-seq library receives an unidentical assortment of RNA fragments, which causes discrepancies in the RT termination events at 5′ of the RNA fragments between libraries.](/cms/asset/215c47b8-21af-447c-b145-bc37dfafd4c1/krnb_a_1740470_f0003_c.jpg)
Figure 4. Novel rG4-seq analysis workflow enables reliable, replicate-free RTS site detection. (A,B) RSR peaks identified ab initio by applying the minimum ΔRSR metric scheme, sequence-based filtering scheme and fragmentation-associated noise model. (A) RSR peaks detected from replicate 1 of HeLa rG4-seq dataset (pairwise comparison between K+/Li+ conditions). (B) RSR peaks detected from replicate 1 of HeLa rG4-seq dataset (pairwise comparison between K+-PDS/Li+ condition). (C,D) Summary of RTS sites detected each replicate rG4-seq dataset, categorized by associated rG4 structural motifs for each site. Adjacent, consecutive RSR peaks were merged and considered to indicate a single RTS site. RTS site detection results (combined analysis of four replicates) from Kwok et al. [Citation9] were included as a reference. (C) RTS sites detected from pairwise comparison between datasets of K+/Li+ conditions (D) RTS sites detected from pairwise comparison between datasets of K+-PDS/Li+ conditions.
![Figure 4. Novel rG4-seq analysis workflow enables reliable, replicate-free RTS site detection. (A,B) RSR peaks identified ab initio by applying the minimum ΔRSR metric scheme, sequence-based filtering scheme and fragmentation-associated noise model. (A) RSR peaks detected from replicate 1 of HeLa rG4-seq dataset (pairwise comparison between K+/Li+ conditions). (B) RSR peaks detected from replicate 1 of HeLa rG4-seq dataset (pairwise comparison between K+-PDS/Li+ condition). (C,D) Summary of RTS sites detected each replicate rG4-seq dataset, categorized by associated rG4 structural motifs for each site. Adjacent, consecutive RSR peaks were merged and considered to indicate a single RTS site. RTS site detection results (combined analysis of four replicates) from Kwok et al. [Citation9] were included as a reference. (C) RTS sites detected from pairwise comparison between datasets of K+/Li+ conditions (D) RTS sites detected from pairwise comparison between datasets of K+-PDS/Li+ conditions.](/cms/asset/c28237b0-c656-45b0-816e-1e56712308a9/krnb_a_1740470_f0004_c.jpg)
Figure 5. Workflow of rG4-seeker pipeline on published HeLa rG4-seq dataset. rG4-seeker is a novel pipeline for rG4-seq analysis that automates the proposed workflow of RTS site detection (minimum ΔRSR metric scheme, sequence-based filtering scheme and fragmentation-associated noise model) and combines detection results from multiple rG4-seq replicate experiments by consensus.
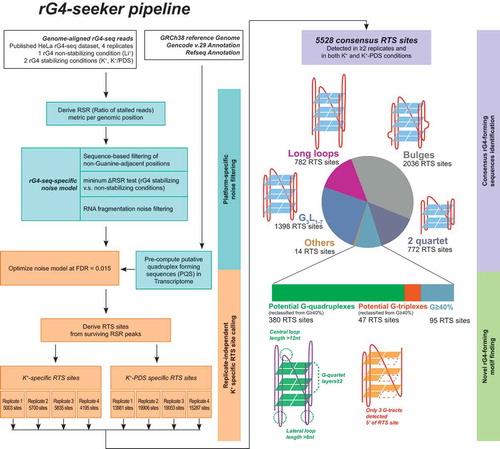
Figure 6. Detected potential G-quadruplexes and G-triplexes suggest novel patterns of structural imperfection toleration in rG4 motifs. (A) Summary of six potential G-quadruplex (rG4)/G-triplex (rG3) candidates selected for RTS PAGE validation. Location of RTS sites revealed by rG4-seq and RTS PAGE assay were indicated separately (* represents one nucleotide position where a RTS event is detected). Most RTS sites discovered by rG4-seq were reproduced in the RTS PAGE assay. (B) RTS PAGE assay of a potential rG4 candidate on MAGOHB gene indicated clear RTS events at the 3′ end of the 3rd and 4th G-tracts, respectively. (C) A potential rG3 was initially suggested to form on DAG1 gene based on findings from rG4-seq. RTS PAGE assay revealed a 4th G-tract 27 nt downstream of the 3rd G-tract, indicating that the region harboured a rG4 motif instead. (D,E) Summary of G-tract and loop lengths of 380 potential G-quadruplexes. The results suggest that toleration for structural imperfection (less than 3 Gs in G-tracts, and longer loops) was higher in the 5′ region and lower at the 3′ end of rG4 motifs. (F,G) Summary of 3′ downstream G-tracts of 47 potential G-triplexes identified by nucleotide sequence searching. 3′ downstream G-tracts were identified in most (43 out of 47) of the rG3s, where most G-tracts were composed of 2 guanine residues.
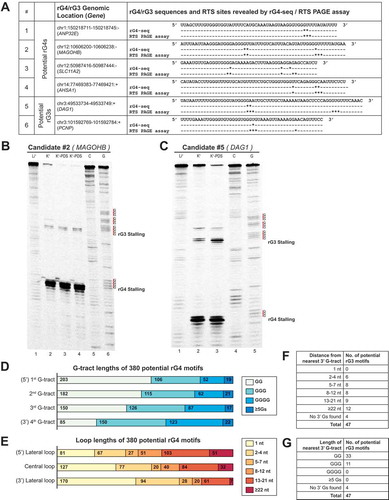
Figure 7. rG4 induces RTS effect in stochastic manner and contributes to divergence between replicates. (A) Breakdown of 5,528 detected consensus RTS sites by their reproducibility in K+ and K+-PDS replicate datasets. Only ~40% of RTS sites were simultaneously detected in all four replicates and in both K+ and K+-PDS conditions. (B) Distribution of 5,528 consensus RTS sites by average read coverage (logarithmic scale) and detection status at the K+ condition. While RTS sites of higher read coverage in general exhibited higher reproducibility in rG4-seq (K+), the marginal improvement in reproducibility by increasing read coverage beyond 32x was subtle. (C) Comparison of rG4-seeker analysis results using only a subset of two or three replicate datasets among four available replicates. Reduced number of replicates caused fewer RTS sites detection but did not increase the detection of RTS sites in the ‘Others’ category (false-positive detections).
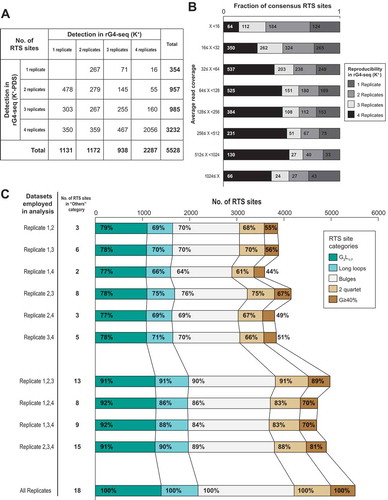
Figure 8. High local GC% may compromise RTS site detection with rG4-seq. (A) Recall statistics of RTS sites (K+) from canonical and non-canonical rG4 categories reported by Kwok et al. [Citation9]. Around 20–25% of RTS sites were not recalled by rG4-seeker in the re-analysis. (B,C) Average GC% (10 nt sliding windows) with shaded standard deviation in the ±100 nt region of the 3,358 RTS sites (K+) reported by Kwok et al. [Citation9], segregated based on their motif categories (canonical or non-canonical) and recall status by rG4-seeker. RTS sites that were not recalled had higher GC% in the 5ʹ and 3ʹ flanking regions when compared with recalled RTS sites.
![Figure 8. High local GC% may compromise RTS site detection with rG4-seq. (A) Recall statistics of RTS sites (K+) from canonical and non-canonical rG4 categories reported by Kwok et al. [Citation9]. Around 20–25% of RTS sites were not recalled by rG4-seeker in the re-analysis. (B,C) Average GC% (10 nt sliding windows) with shaded standard deviation in the ±100 nt region of the 3,358 RTS sites (K+) reported by Kwok et al. [Citation9], segregated based on their motif categories (canonical or non-canonical) and recall status by rG4-seeker. RTS sites that were not recalled had higher GC% in the 5ʹ and 3ʹ flanking regions when compared with recalled RTS sites.](/cms/asset/bb13e5de-3509-4e49-bbe2-bbfde2b4e685/krnb_a_1740470_f0008_c.jpg)
Supplemental Material
Download Zip (18.8 MB)Availability
rG4-seeker is open source software available in the GitHub repository (https://github.com/TF-Chan-Lab/rG4-seeker)
