Figures & data
Figure 1. LINC01198 was up-regulated in GBM. (A–C) LINC01198 expression profiles in human normal tissues from UCSC, NCBI and NONCODE. (D) LINC01198 expression pattern in different cancer types. Data were obtained from TCGA. (E, F) LINC01198 expression in GBM tissues and its association with GBM prognosis were all obtained from TCGA database.
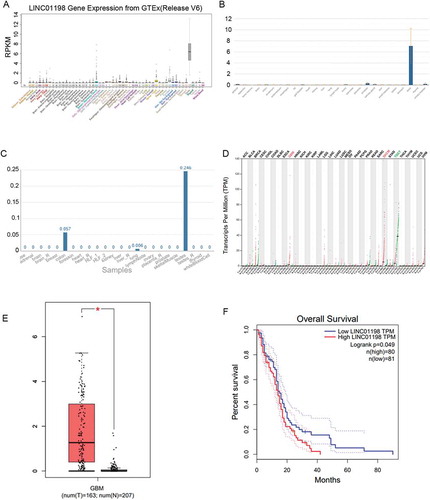
Figure 2. LINC01198 was highly expressed and promoted cell proliferation in glioma. (A) qRT-PCR results showed that LINC01198 was up-regulated in glioma cells. (B) The transfection efficiency of shLINC01198#1 or shLINC01198#2 was assayed in U87 and U251 via qRT-PCR. (C–F) MTT, EdU (scale bar = 100 μm), colony formation and TUNEL (scale bar = 100 μm) assays were performed to detect proliferative and apoptotic cells after silencing LINC01198 expression by shLINC01198#1/2. (G) Apoptosis-related proteins (Bcl-2, Bax and Caspase 3) were examined by Western blot in U87 and U251 cells with or without LINC01198 inhibition. **P < 0.01.
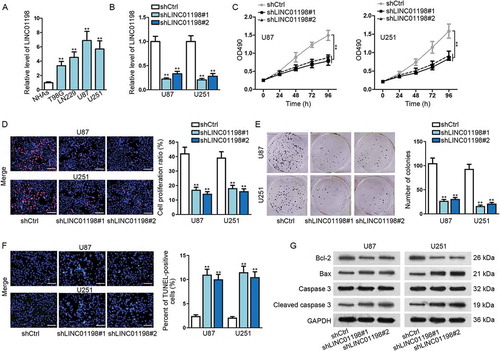
Figure 3. LINC01198 activated PI3 K/AKT signalling pathway in glioma. (A, B) The protein levels of AKT, p-AKT (T308), mTOR and p-mTOR in glioma cells under indicated transfections were tested by Western blotting. (C) The effect of SC79 on shLINC01198#1 or shLINC01198#2-affected glioma cell proliferation and apoptosis was measured by MTT, EdU (scale bar = 100 μm), colony formation, TUNEL (scale bar = 100 μm) and Western blot assays in both U87 and U251 cells. **P < 0.01.
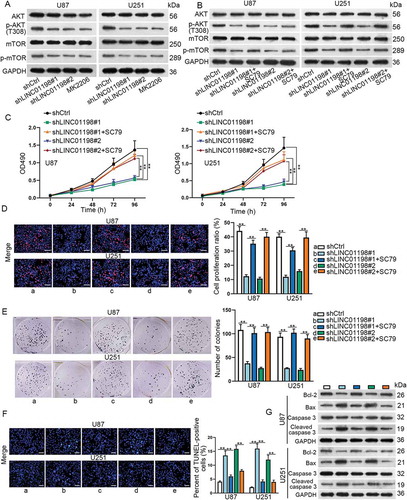
Figure 4. LINC01198 enhanced PIK3CA expression by sponging miR-129-5p in glioma. (A) The distribution of LINC01198 in glioma cells was examined by FISH assay (scale bar = 50 μm). (B–D) Ago2-RIP, RNA pull-down and luciferase reporter assays were conducted for evaluating the interaction between miR-129-5p and LINC01198 or PIK3CA in glioma cells. (E) The expression of PIK3CA in indicated U251 and U87 cells was measured by qRT-PCR. (F–G) The levels of LINC01198 and PIK3CA in U251 and U87 cells with overexpression of full-length or mutant LINC01198 were analysed by qRT-PCR. (H) RNA pull-down assay unveiled a decline of miR-129-5p-targeted PIK3CA mRNA upon up-regulation of full-length LINC01198 rather than mutated LINC01198. *P < 0.05, **P < 0.01, n.s. meant no significance.
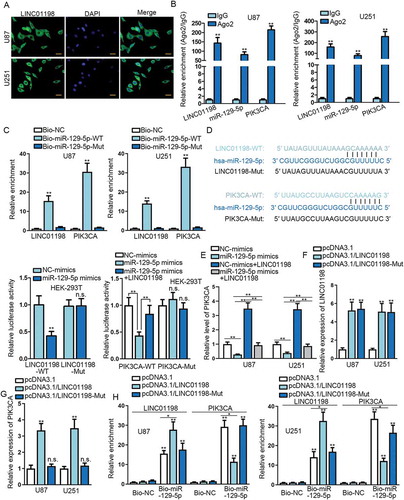
Figure 5. PTEN transcription was inhibited by the REST/RCOR1/HDAC2 complex in glioma. (A–C) Transfection efficiency of pcDNA3.1/REST, pcDNA3.1/RCOR1 or pcDNA3.1/HDAC2 in U87 and U251 cells were assayed via Western blot. Relative expression of PTEN in U87 and U251 cells transfected with above transfection plasmids was determined by qRT-PCR. (D–G) The inhibition of REST/RCOR1/HDAC2 complex on PTEN transcription was verified through ChIP and luciferase reporter assays. **P < 0.01.
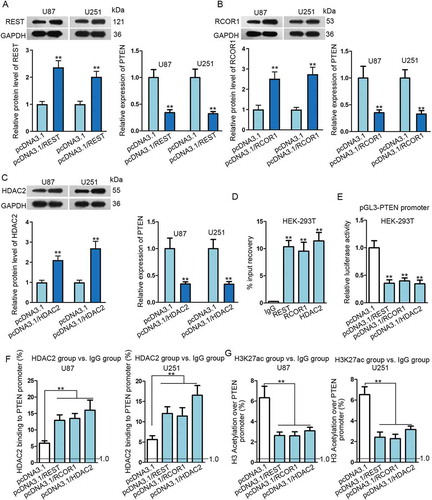
Figure 6. LINC01198 promoted the formation of REST/RCOR1/HDAC2 complex in glioma. (A–C) The interaction of LINC01198 with REST, RCOR1 or HDAC2 was explored via RIP assay, RNA pull-down and Co-IP assay. (D) ChIP assay was carried out to evaluate the impact of LINC01198 silence on the binding of REST, RCOR1 and HDAC2 to PTEN promoter. RNA pol Ⅱ was the negative control. (E) Influence of LINC01198 inhibition on histone acetylation of PTEN promoter was assessed by ChIP assays. Promoters of MINPP1 and RNLS were negative controls for PTEN promoter. **P < 0.01, n.s. meant no significance.
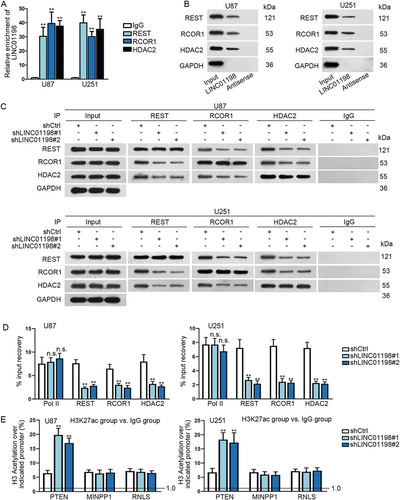
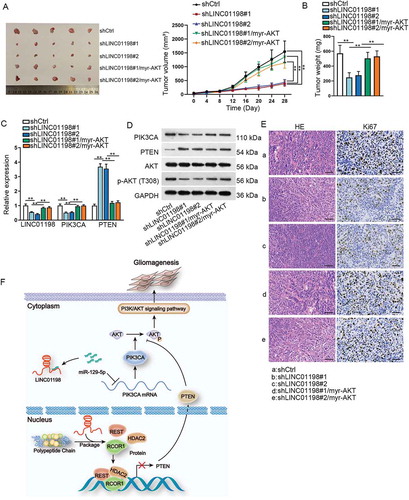
Figure 7. LINC01198 promoted glioma tumour growth in vivo through AKT-dependent pathway. (A) Representative images and the corresponding growth curves of tumours from mice injected with indicated U87 cells. (B) Weight of abovementioned tumours. (C–D) The expressions of LINC01198, PIK3CA and PTEN, as well as protein levels of PIK3CA, PTEN, AKT and p-AKT (T308) in these xenografts, were detected by qRT-PCR and Western blot, respectively. (E) Tumour histology was studied by HE staining (scale bar = 200 μm), while the proliferation marker Ki67 was estimated by IHC staining (scale bar = 200 μm). (F) Schedule model of the regulatory mechanism of LINC01198 in gliomagenesis. **P < 0.01.