Figures & data
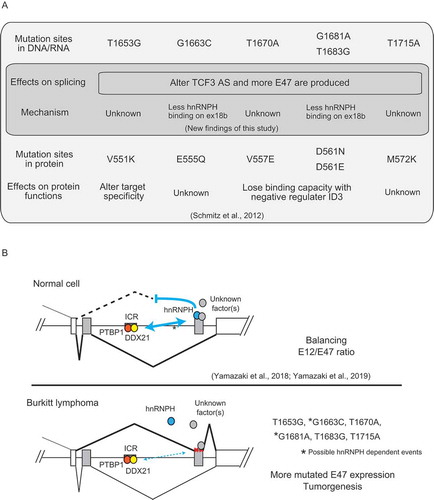
Figure 1. Recurrent mutations of the TCF3 gene in BL affect TCF3 alternative splicing.
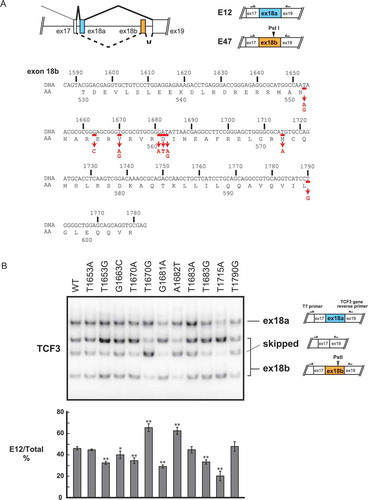
Figure 2. Isoform-specific regulation of TCF3’s downstream target genes PTPN6 and CCND3.
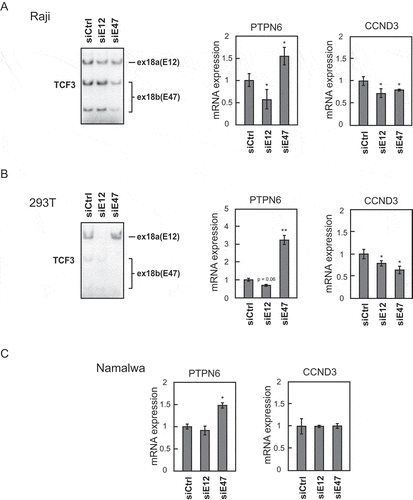
Figure 3. Recurrent TCF3 mutations can disturb hnRNPH1 binding to exon 18b.
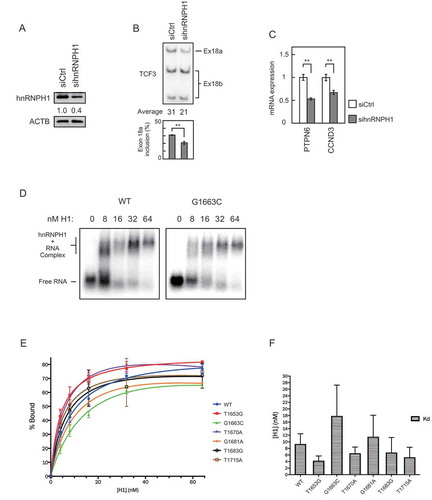
Figure 4. Summary of the effects of recurrent TCF3 mutations in BL, and a model of their effects on TCF3 AS regulation.