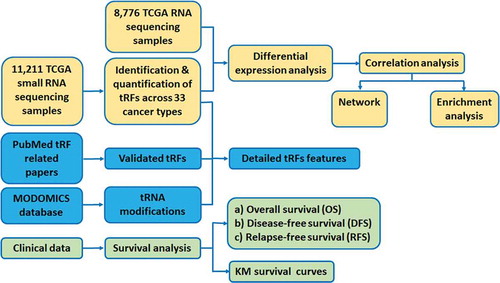
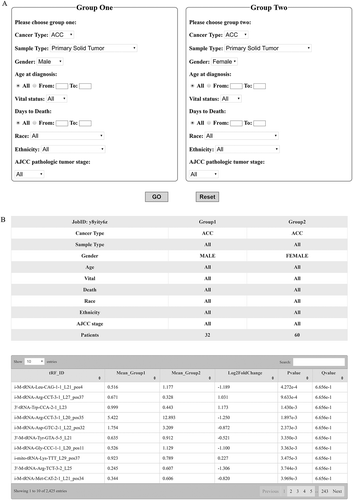
Figures & data
Table 1. Summary of TCGA samples used in the study and 5ʹ-tRFs, 3ʹ-tRFs,3ʹU-tRFs and i-tRFs identified in the study.
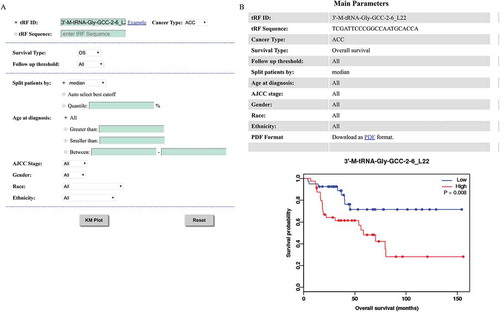
Figure 2. Search functions of OncotRF. (A) Search result of 3ʹ-M-tRNA-Gly-GCC-2-6_L22. (B) 3ʹ-M-tRNA-Gly-GCC-2-6_L22 expression in cancers. (C) Validation result of 3ʹ-M-tRNA-Gly-GCC-2-6_L22. (D) 3ʹ-M-tRNA-Gly-GCC-2-6_L22 alignment with tRNA-Gly-GCC-2-1, its position on the secondary structure of tRNA-Gly-GCC-2-1, and possible modifications of tRNA-Gly-GCC-2-1 from MODIFICS database.
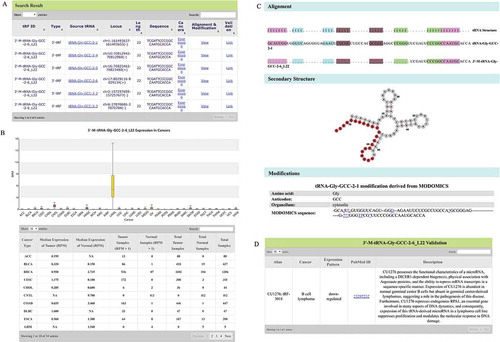
Figure 3. Cancer functions. (A) Differentially expressed 3ʹU-tRFs in BLCA. (B) Differentially expressed mRNAs in BLCA. (C) Correlation analysis of tRFs and mRNAs in BLCA. tRF-mRNA pairs with their absolute correlation coefficients > 0.4 (i.e., |r|>0.4) were presented. (D) Network analysis of differentially expressed tRFs and mRNAs in BLCA. tRF-mRNA pairs with |r|>0.4 in Figure 3 C were subjected to network analysis. (E) Functional enrichment analysis of genes that are co-expressed with 3ʹ- U-tRFs (|r|>0.4). (F) Survival analysis of differentially expressed 3ʹU-tRFs in BLCA.