Figures & data
Figure 1. Identification of lncRNAs transcriptionally induced/repressed by oestrogen signalling in ER+ breast cancer cells.
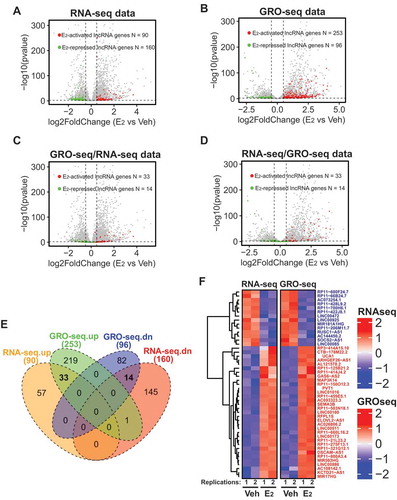
Figure 2. Expression of oestrogen-regulated lncRNAs correlates with eRNA production of neighbouring ERα enhancers.
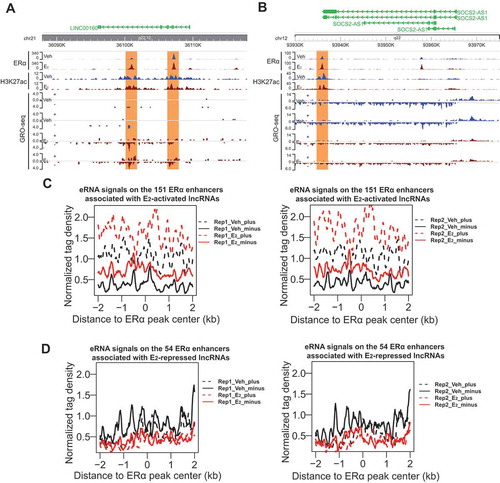
Figure 3. Transcription of oestrogen-regulated lncRNAs is associated with the presence of enhancer complex.
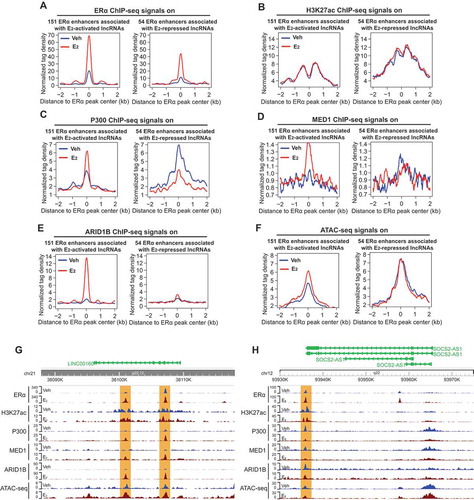
Figure 4. LINC00160 and RUNX1 are co-regulated by E2 and are located within one TAD.
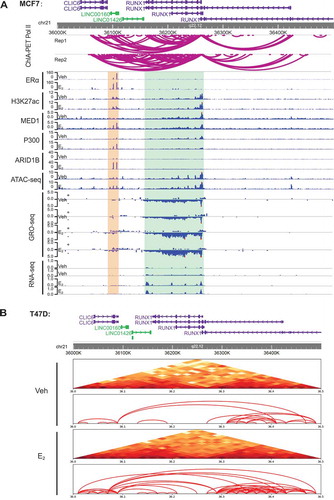
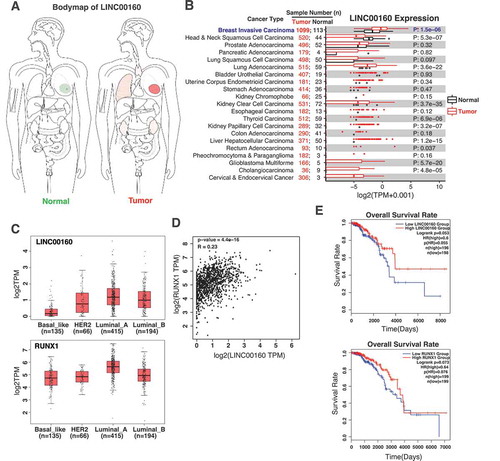
Figure 5. The levels of LINC00160 and RUNX1 correlate with the disease outcome in ER+ breast cancer patients.