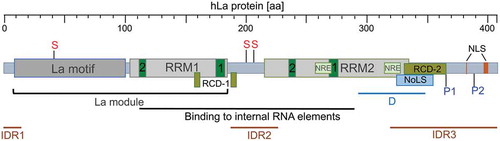
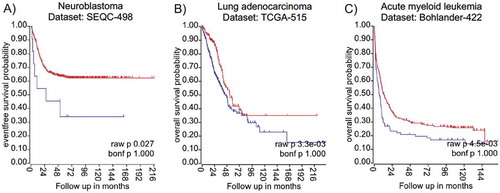
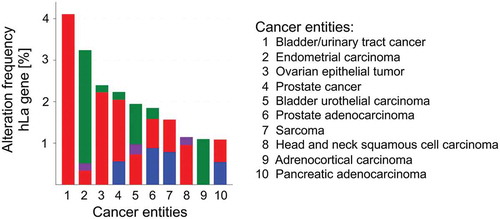
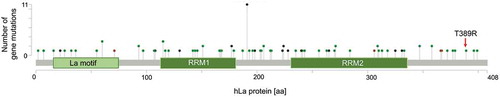
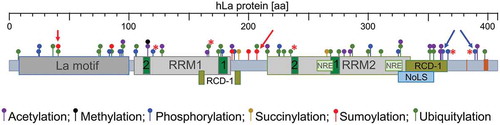
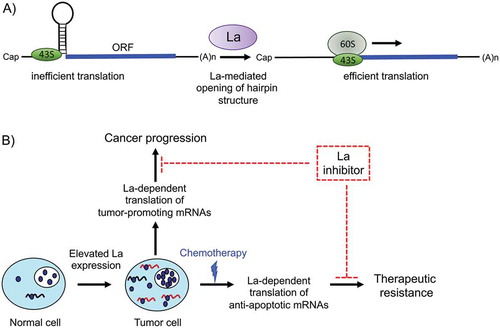
Figures & data
Kuehnert J, Sommer G, Zierk AW, et al. Novel RNA chaperone domain of RNA-binding protein La is regulated by AKT phosphorylation. Nucleic Acids Res. 2015;43:581–594. Hussain RH, Zawawi M, Bayfield MA. Conservation of RNA chaperone activity of the human La-related proteins 4, 6 and 7. Nucleic Acids Res. 2013;41:8715–8725. Naeeni AR, Conte MR, Bayfield MA. RNA chaperone activity of human La protein is mediated by variant RNA recognition motif. J Biol Chem. 2012;287:5472–5482. Simons FH, Broers FJ, Van Venrooij WJ, et al. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp Cell Res. 1996;224:224–236. Bayfield MA, Kaiser TE, Intine RV, et al. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol Cell Biol. 2007;27:3303–3312. Intine RV, Dundr M, Misteli T, et al. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol Cell. 2002;9:1113–1123. Horke S, Reumann K, Schweizer M, et al. Nuclear trafficking of La protein depends on a newly identified nucleolar localization signal and the ability to bind RNA. J Biol Chem. 2004;279:26563–26570. Epub 2004 Apr 1. Intine RV, Dundr M, Vassilev A, et al. Nonphosphorylated human La antigen interacts with nucleolin at nucleolar sites involved in rRNA biogenesis. Mol Cell Biol. 2004;24:10894–10904. Craig A, Svitkin Y, Lee H, et al. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol Cell Biol. 1997;17:163–169. Kucera NJ, Hodsdon ME, Wolin SL. An intrinsically disordered C terminus allows the La protein to assist the biogenesis of diverse noncoding RNA precursors. Proc Natl Acad Sci U S A. 2011;108:1308–1313. van Niekerk EA, Willis DE, Chang JH, et al. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc Natl Acad Sci U S A. 2007;104:12913–12918. Kota V, Sommer G, Hazard ES, et al. SUMO modification of the RNA-binding protein La regulates cell proliferation and STAT3 protein stability. Mol Cell Biol. 2018;38(2):e00129-17. Brenet F, Socci ND, Sonenberg N, Holland EC. Akt phosphorylation of La regulates specific mRNA translation in glial progenitors. Oncogene. 2009;28:128–139. Broekhuis CH, Neubauer G, van Der Heijden A, et al. Detailed analysis of the phosphorylation of the human La (SS-B) autoantigen. (De)phosphorylation does not affect its subcellular distribution. Biochemistry. 2000;39:3023–3033. Fan H, Sakulich AL, Goodier JL, et al. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715.