Figures & data
Figure 1. Canonical and non-canonical splicing and their products. A single gene is able to be canonically and non-canonically spliced (backsplicing) to generate mRNAs and regular RNA circles (ecircRNA, ciRNA and EIcircRNA). Circularization process is mediated by base-pairing between flanking introns and by some RBPs
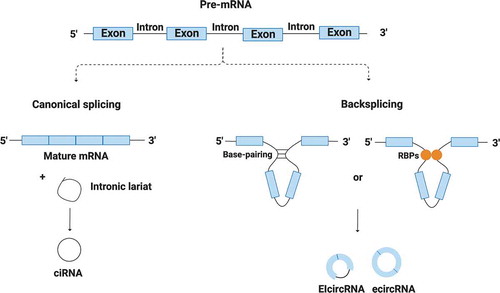
Figure 2. Biogenesis of rt-circRNAs. Read-through transcription results in the formation of hybrid circRNAs (rt-circRNAs) that include coding exons from two adjacent and similarly oriented genes. Circularization process is mediated by base-pairing between long introns that harbour repetitive sequences
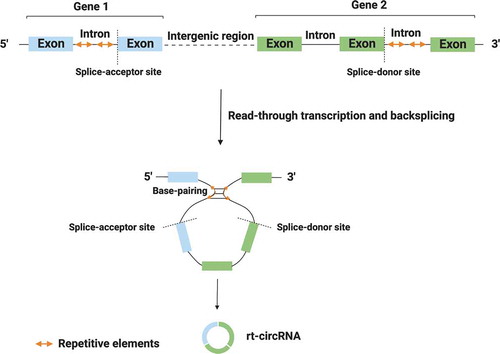
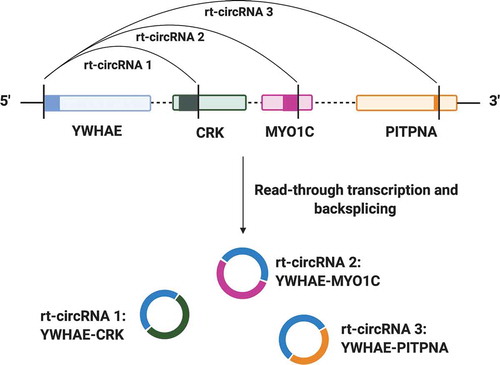
Figure 3. Example of the biogenesis of rt-circRNAs by read-through transcription. YWHAE produces three rt-circRNAs with three different partner genes (CRK, MYO1C and PITPNA), demonstrating the plasticity of RNA processing mechanisms. Solid coloured blocks represent exons. Vertical black lines represent splice sites