Figures & data
Figure 1. DDX55 is present in the nucleoplasm and associates with late pre-LSU complexes. (A) The sub-cellular localization of Flag-tagged DDX55 was determined by immunofluorescence (DDX55) using antibodies against the Flag tag. Co-immunofluorescence using an α-UTP14A served as a nucleolar marker and nuclear material was visualized using DAPI staining. Scale bar represents 10 μm. (B) Whole cell extracts prepared from HEK293 cells expressing DDX55-Flag were separated by sucrose density gradient centrifugation. During fractionation, a profile of absorbance at 260 nm was generated and the peaks corresponding to free proteins/small complexes, 80S mature/90S pre-ribosomes, and (pre-)40S and (pre-)60 particles are indicated (upper panel). Proteins in each fraction were separated by SDS-PAGE and analysed by western blotting using an α-Flag antibody (lower panel)
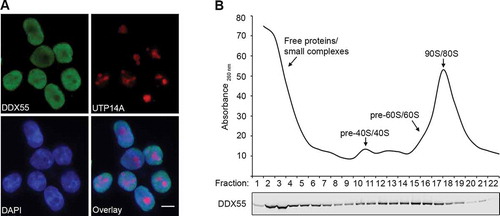
Figure 2. DDX55 crosslinks to helices H62-63 of the 28S ribosomal RNA sequence in vivo. (A) HEK293 cells expressing DDX55-Flag or the Flag tag alone were crosslinked using light at 254 nm. The tagged proteins and crosslinked RNAs were retrieved on α-Flag beads under native conditions and subjected to a partial RNase digest. Complexes were then immobilized on NiNTA in denaturing conditions and co-purified RNA fragments were [32P] labelled, ligated to sequencing adaptors. After elution, protein-RNA complexes were separated by denaturing PAGE, transferred to a nitrocellulose membrane and radiolabelled RNAs were detected by autoradiography. (B) The regions of the membranes containing crosslinked DDX55-Flag complexes were excised, as well as a corresponding region of the lane containing the Flag sample. RNAs were released by proteinase treatment, purified and converted to a cDNA library that was subjected to deep sequencing. The obtained sequence reads were mapped to the human genome and the relative proportions of reads mapped to gene features encoding different classes of RNA was determined. Abbreviations – ribosomal RNA (rRNA), transfer RNA (tRNA), small nucleolar RNA (snoRNA), non-coding RNA (ncRNA), small nuclear RNA (snRNA), long non-coding RNA (lncRNA), microRNA (miRNA), mitochondrial RNA (mtRNA). (C) The number of reads mapping to each nucleotide of the rDNA sequence encoding the 47S pre-rRNA is shown above a schematic view of the transcript (upper panel). The number of mutations, arising during reverse transcription due to the presence of amino acid-crosslinked nucleotides, mapping to each nucleotide is also shown (lower panel). (D) The number of sequencing reads in the DDX55-His6-Prc-Flag CRAC data mapping to each nucleotide of the 28S rRNA sequence is shown on the secondary structure of the mature 28S rRNA using the indicated colour scale. A magnified view of the region crosslinked by DDX55 is shown. (E) The number of sequencing reads in the DDX55-His6-Prc-Flag CRAC data mapping to each nucleotide of the tertiary structure of the 28S rRNA (grey) within the mature LSU (PBD: 4V6X) is shown using a colour scale as in (D). The ribosomal proteins of the LSU are shown in surface view in pale cyan. The positions of key ribosomal features are indicated. CP – central protuberance
![Figure 2. DDX55 crosslinks to helices H62-63 of the 28S ribosomal RNA sequence in vivo. (A) HEK293 cells expressing DDX55-Flag or the Flag tag alone were crosslinked using light at 254 nm. The tagged proteins and crosslinked RNAs were retrieved on α-Flag beads under native conditions and subjected to a partial RNase digest. Complexes were then immobilized on NiNTA in denaturing conditions and co-purified RNA fragments were [32P] labelled, ligated to sequencing adaptors. After elution, protein-RNA complexes were separated by denaturing PAGE, transferred to a nitrocellulose membrane and radiolabelled RNAs were detected by autoradiography. (B) The regions of the membranes containing crosslinked DDX55-Flag complexes were excised, as well as a corresponding region of the lane containing the Flag sample. RNAs were released by proteinase treatment, purified and converted to a cDNA library that was subjected to deep sequencing. The obtained sequence reads were mapped to the human genome and the relative proportions of reads mapped to gene features encoding different classes of RNA was determined. Abbreviations – ribosomal RNA (rRNA), transfer RNA (tRNA), small nucleolar RNA (snoRNA), non-coding RNA (ncRNA), small nuclear RNA (snRNA), long non-coding RNA (lncRNA), microRNA (miRNA), mitochondrial RNA (mtRNA). (C) The number of reads mapping to each nucleotide of the rDNA sequence encoding the 47S pre-rRNA is shown above a schematic view of the transcript (upper panel). The number of mutations, arising during reverse transcription due to the presence of amino acid-crosslinked nucleotides, mapping to each nucleotide is also shown (lower panel). (D) The number of sequencing reads in the DDX55-His6-Prc-Flag CRAC data mapping to each nucleotide of the 28S rRNA sequence is shown on the secondary structure of the mature 28S rRNA using the indicated colour scale. A magnified view of the region crosslinked by DDX55 is shown. (E) The number of sequencing reads in the DDX55-His6-Prc-Flag CRAC data mapping to each nucleotide of the tertiary structure of the 28S rRNA (grey) within the mature LSU (PBD: 4V6X) is shown using a colour scale as in (D). The ribosomal proteins of the LSU are shown in surface view in pale cyan. The positions of key ribosomal features are indicated. CP – central protuberance](/cms/asset/79189e36-3f40-42db-b959-328b209b1ea8/krnb_a_1829366_f0002_c.jpg)
Figure 3. Pre-rRNA processing in cells depleted of DDX55. (A) Simplified processing scheme showing the major pre-rRNA intermediates present in human cells. Mature rRNA sequences are shown as black rectangles, and internal and external transcribed spacers (ITS and ETS respectively) are represented by black lines. The hybridization positions of probes used for northern blotting are indicated by triangles on the initial 47S pre-rRNA transcript. (B) HEK293 cells expressing the Flag tag or siRNA-insensitive, Flag-tagged DDX55 (DDX55siins-Flag) were treated with siRNAs targeting the firefly luciferase (siNT) or DDX55 (si55). Cells were harvested 90 h after transfection, and proteins were separated by SDS-PAGE and analysed by western blot using antibodies against DDX55 (α-DDX55) or, as a loading control tubulin (α-tubulin). (C) Total RNA prepared from siRNA-treated cells as in (B) was separated by denaturing agarose gel electrophoresis and transferred to a nylon membrane. Pre-rRNA species and the actin mRNA were detected by northern blotting using the probes indicated to the right of the panel. (D) The levels of pre-rRNA species in three independent experiments were quantified, normalized according to the actin mRNA, and are presented relative to the Flag, siNT sample as mean ± standard error. (E) HeLa cells treated with siNT or siDDX55 were subjected to pulse-chase metabolic labelling. Total RNAs were separated by denaturing agarose gel electrophoresis and transferred to a nylon membrane. Abundant labelled RNAs were detected using a phosphorimager. (F) The levels of (pre-)rRNA species in three independent pulse-chase experiments were quantified, normalized according to the 18S rRNA, and are presented relative to the siNT sample as mean ± standard error
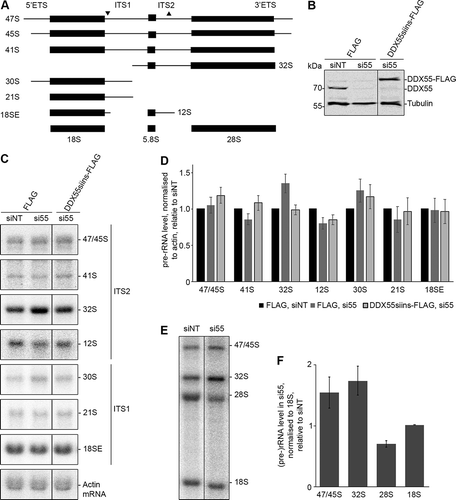
Figure 4. The C-terminal region of DDX55 contains a nuclear localization signal. (A) Schematic overview of DDX55. Amino acid numbers corresponding to specific domain boundaries (www.uniprot.org) are given above. (B) The amino acid sequences of human DDX55 and its homologues from the species indicated were aligned. Basic amino acids are highlighted in red, evolutionarily conserved amino acids are marked with a black background, and conserved basic amino acids are shown in white with a red background. A putative bipartite nuclear localization signal (NLS) identified in the human DDX55 sequence is indicated by a green box. (C) The subcellular localization of Flag tagged, full-length DDX55 (DDX55), DDX55 lacking the C-terminal tail (DDX551-403), DDX55 lacking the C-terminal tail but coupled to the NLS of the SV40 large T-antigen (DDX551-403+NLS) and DDX55 carrying amino acid substitutions (K537A, R538A, K539A, R540A) within the predicted NLS sequence (DDX55NLSmut-Flag) were determined by immunofluorescence using an α-Flag antibody. Co-immunofluorescence using α-UTP14A served as a nucleolar marker and nuclear material was visualized using DAPI staining. Scale bar represents 10 μm. (D) The subcellular localization of a GFP-GFP-GST reporter (GGG) and the reporter coupled to the predicted DDX55 NLS (GGG-DDX55533-563) was determined by fluorescence microscopy. DAPI staining was used to visualize nuclear material. Scale bar represents 10 μm
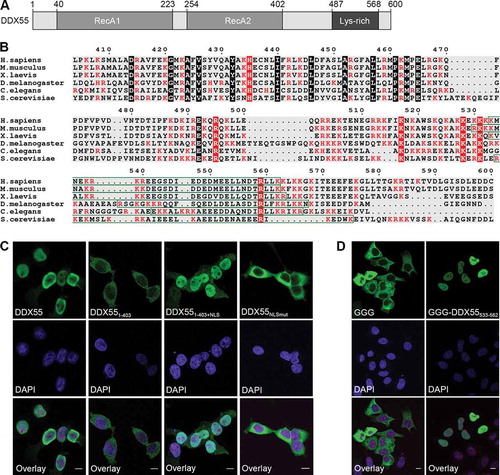
Figure 5. The ATPase DDX55 preferentially binds and is stimulated by double-stranded RNAs. (A) ZZ-DDX55-His7, ZZ-DDX551-403-His7 and ZZ-DDX55T60A-His7 were recombinantly expressed in E. coli and purified via their His tags on a nickel matrix. Purified proteins were separated by SDS-PAGE and visualized by Coomassie staining. (B,C) Fluorescence anisotropy experiments were performed using fluorescently labelled, single-stranded (B) or double-stranded (C) RNAs and different amounts of purified ZZ-DDX55-His7 and ZZ-DDX551-403-His7. Data from three independent experiments are presented as mean ± standard deviation. (D) In vitro NADH-coupled ATPase assays were performed using no protein (no prot.), DDX55, DDX55T60A or DDX551-403. Samples contained no RNA (-), single- or double-stranded RNA (ss and ds respectively) or a mimic of the cellular RNA sequence crosslinked by DDX55 (H62)
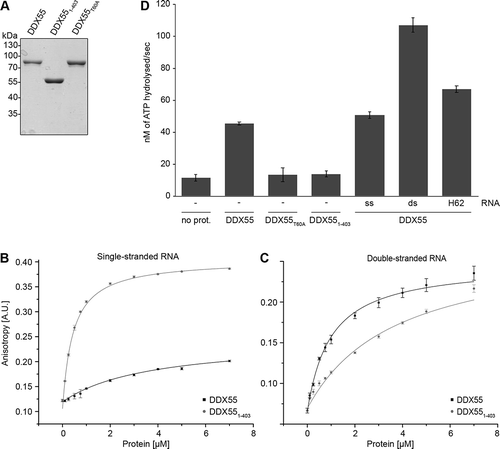
Figure 6. The C-terminal region of DDX55 is required for pre-ribosome association. (A) HEK293 cells expressing the Flag tag alone, DDX55-Flag, DDX551-403-Flag or DDX551-403+NLS-Flag were crosslinked using light at 254 nm. The tagged protein and crosslinked RNAs were retrieved on α-Flag beads under native conditions and subjected to a partial RNase digest. Complexes were then immobilized on NiNTA in denaturing conditions and co-purified RNA fragments were [32P] labelled. After elution, protein-RNA complexes were separated by denaturing PAGE, transferred to a nitrocellulose membrane and radiolabelled RNAs were detected by autoradiography. Protein samples were separated by SDS-PAGE and subjected to western blotting using an α-Flag antibody. (B) Whole cell extracts prepared from HEK293 cells expressing HEK293 cells expressing DDX55-Flag, DDX551-403-Flag or DDX551-403+NLS-Flag were separated by sucrose density gradient centrifugation. During fractionation, an absorbance profile at 260 nm was generated and the peaks corresponding to 80S mature/90S pre-ribosomes, and (pre-)40S and (pre-)60 particles are indicated (upper panel). Proteins in each fraction were separated by SDS-PAGE and analysed by western blotting using an α-Flag antibody (lower panel)
![Figure 6. The C-terminal region of DDX55 is required for pre-ribosome association. (A) HEK293 cells expressing the Flag tag alone, DDX55-Flag, DDX551-403-Flag or DDX551-403+NLS-Flag were crosslinked using light at 254 nm. The tagged protein and crosslinked RNAs were retrieved on α-Flag beads under native conditions and subjected to a partial RNase digest. Complexes were then immobilized on NiNTA in denaturing conditions and co-purified RNA fragments were [32P] labelled. After elution, protein-RNA complexes were separated by denaturing PAGE, transferred to a nitrocellulose membrane and radiolabelled RNAs were detected by autoradiography. Protein samples were separated by SDS-PAGE and subjected to western blotting using an α-Flag antibody. (B) Whole cell extracts prepared from HEK293 cells expressing HEK293 cells expressing DDX55-Flag, DDX551-403-Flag or DDX551-403+NLS-Flag were separated by sucrose density gradient centrifugation. During fractionation, an absorbance profile at 260 nm was generated and the peaks corresponding to 80S mature/90S pre-ribosomes, and (pre-)40S and (pre-)60 particles are indicated (upper panel). Proteins in each fraction were separated by SDS-PAGE and analysed by western blotting using an α-Flag antibody (lower panel)](/cms/asset/4fbe66bc-26ad-4aa6-b912-841f10eacff9/krnb_a_1829366_f0006_b.gif)
