Figures & data
Figure 1. (A) Representations of ZIKV, TBEV, MODV and RBV xrRNA structures, illustrating two- (above) and three-dimensional (below) contacts. The proposed stem-interactions α, β, γ, δ and ε are given in blue, green, magenta, red and orange, respectively. Nucleotides involved in the base triple interactions are shown in bold. (B) Sequences of ZIKV and CFAV xrRNA, in structural alignment with a collection of various tick-borne and no-known-vector flaviviruses, revealing covariations in this region. Grey columns in the background depict which nucleotides are involved in the base triple interaction. Accession numbers used for sequences were: Zika virus isolate 15555 (ZIKV), MN025403; Cell fusing agent virus isolate Guadeloupe (LR694081); Tick-borne encephalitis virus Sofjin-HO (TBEV), AB062064; Spanish goat encephalitis virus (SGEV), NC_027709; Tick-borne encephalitis virus Neudoerfl (WEST), U27495; Langat virus strain TP21 (LANG), AF253419; Louping ill virus LI3/1 (LOUP), KP144331; Karshi virus strain LEIV 2247 (KARS), AY863002; Deer tick virus strain ctb30 (DEER), AF311056; Powassan virus strain LB (POWA), L06436; Modoc virus (MODV), NC_003635; Rio Bravo virus (RBV), JQ582840; Montana myotis leukoencephalitis virus (MMLV), NC_004119; Apoi virus (APOIV), AF452050
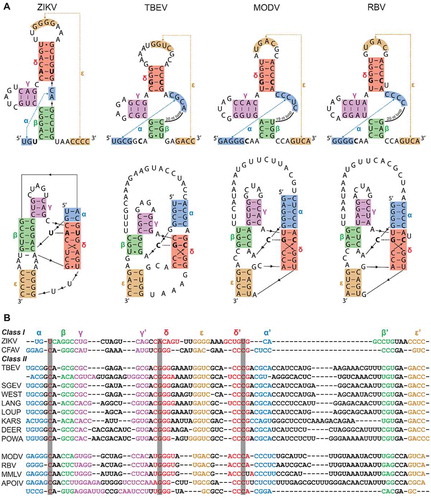
Figure 2. In vitro Xrn1 degradation assays probing (A) the Xrn1-resistance of ‘minimal’ RNA constructs for TBEV, MODV and RBV, and (B) the Xrn1-resistance of these constructs carrying loop deletions. RNAs, incubated with or without Xrn1, were loaded on denaturing polyacrylamide gels shown here with the corresponding names of constructs given above. Data below the gels depict the average percentage (± SD) of Xrn1-resistant RNA
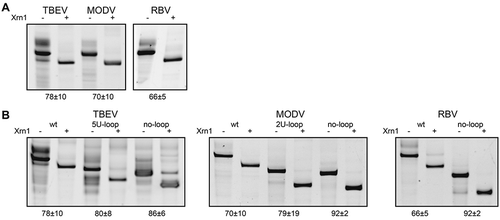
Figure 3. In vitro Xrn1 degradation assays performed a variety of constructs and loaded on denaturing polyacrylamide gels, testing the effect of mutations targeted at the (A) β stem, (B) ε stem and (C) the base triple interaction. Data below the gels depict the average percentage (± SD) of Xrn1-resistant RNA. ‘N.D.’ signifies that this value could not be determined reliably, but does not exceed 10%
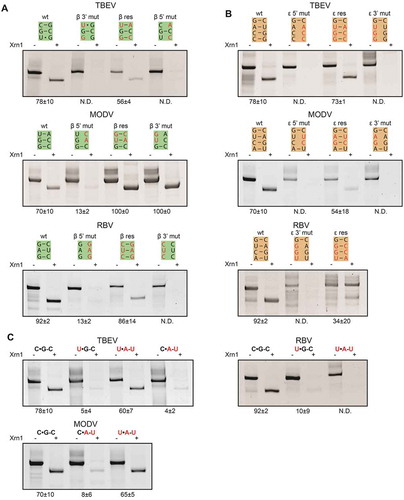
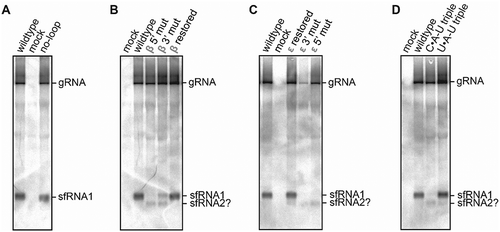
Figure 4. Northern blot analysis of total RNA isolated from BHK-21 cells transfected with RNA derived from infectious MODV cDNA clones. The clone varieties are listed above the lanes, corresponding with the no-loop (A), β stem (B), ε stem (C) and base triple (D) mutants as used in in vitro Xrn1 degradation assays