Figures & data
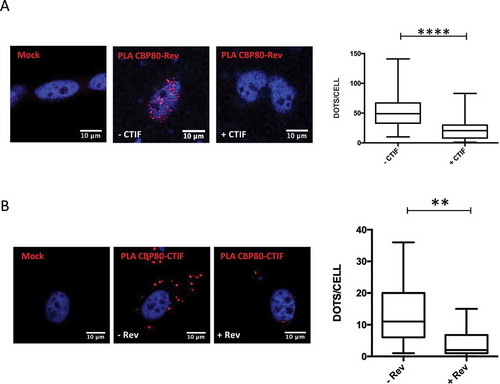
Figure 1. CTIF inhibits HIV-1 and HIV-2 Gag synthesis
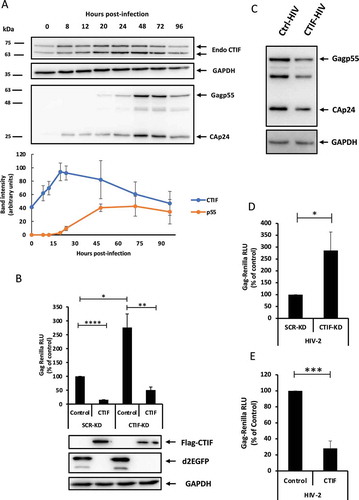
Figure 2. CTIF acts on Rev function
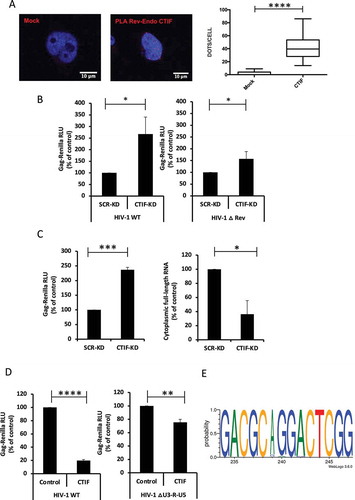
Figure 3. CTIF inhibits Gag synthesis through its N-terminal domain
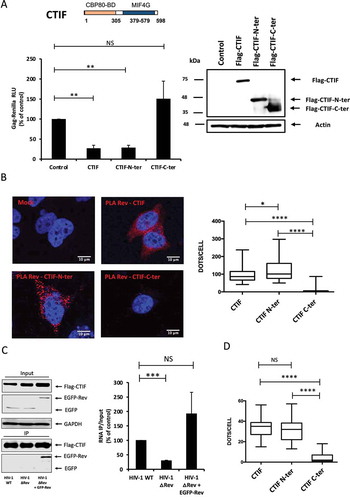
Figure 4. CTIF modifies Rev localization within the cell
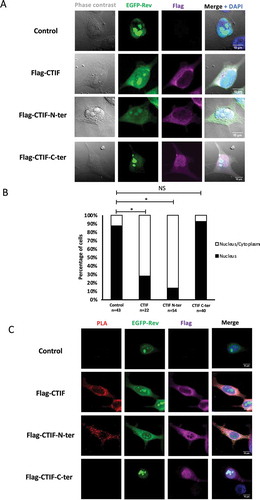
Figure 5. And interferes with the Rev-CBP80 interaction