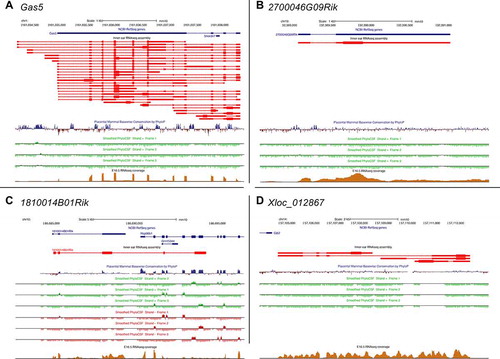
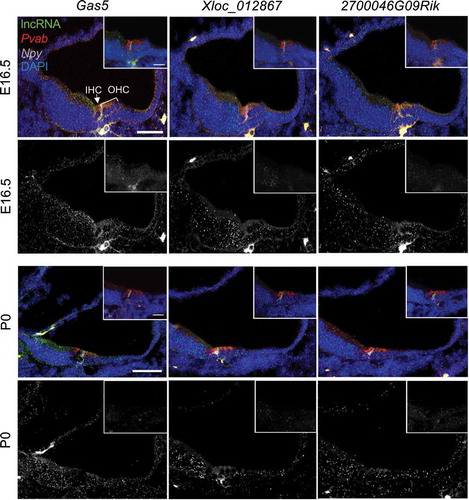
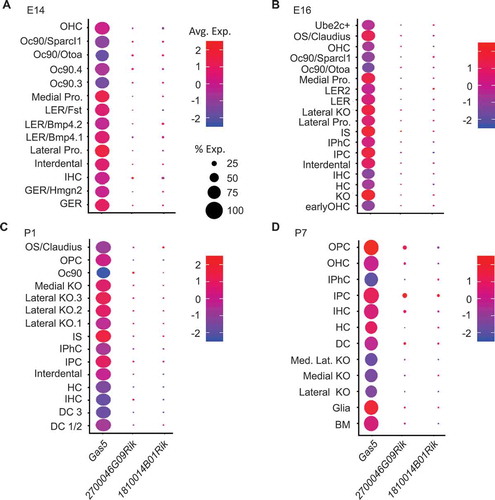
Expression of
Gas5, 2700046G09Rik, and
1810014B01Rik in the indicated cell types based on single cell RNA-seq data [
Citation21] at the indicated developmental time points: (A) E14, (B) E16, (C) P1, and (D) P7. Size of each circle indicates percent of cells for each cell type expressing the indicated lncRNA. Colour code indicates scaled (z-score) average expression in each cell. BM (basement membrane), DC (Deiters’ cells), DC1/2 (Deiters’ cells rows 1 and 2), DC 3 (Deiters’ cells row 3), GER (greater epithelial ridge), GER/Hmgn2 (greater epithelial ridge/Hmgn2 expressing cells), HC (hair cells), IHC (inner hair calls), IPC (inner pillar cells), IPhC (inner phalangeal cells), IS (inner sulcus cells), KO (Kölliker’s organ cells), Lateral KO (lateral Kölliker’s organ cells), Lateral KO.1 (lateral Kölliker’s organ cells type 1), Lateral KO.2 (lateral Kölliker’s organ cells type 2), Lateral KO.3 (lateral Kölliker’s organ cells type 3), Lateral pro. (Lateral prosensory), LER (lesser epithelial ridge), LER2 (lesser epithelial ridge type 2), LER/Bmp4.1 (lesser epithelial ridge/BMP4 type 1 expressing cells), LER/Bmp4.2 (lesser epithelial ridge/BMP4 type 2 expressing cells), LER/Fst (lesser epithelial ridge/Fst expressing cells), Medial KO (medial Kölliker’s organ cells), Med. Lat. KO (medial lateral Kölliker’s organ cells), Medial pro. (Medial prosensory), Oc90 (Otoconin90 expressing cells), Oc90.3 (Otoconin90 type 3 expressing cells), Oc90.4 (Otoconin90 type 4 expressing cells), Oc90/Ota (Otoconin90/Otoa expressing cells), Oc90/Sparcl1 (Otoconin90/Sparcl1 expressing cells), OHC (Outer hair cells), OPC (outer pillar cells), Os/Claudius (outer sulcus cells/Claudius), Ube2c+ (Ube2c expressing cells).