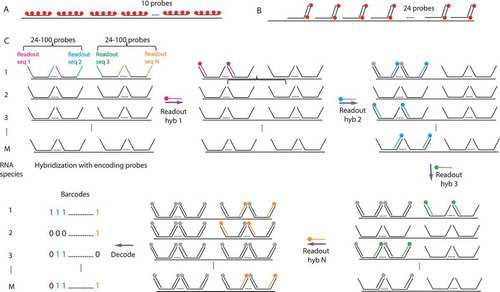
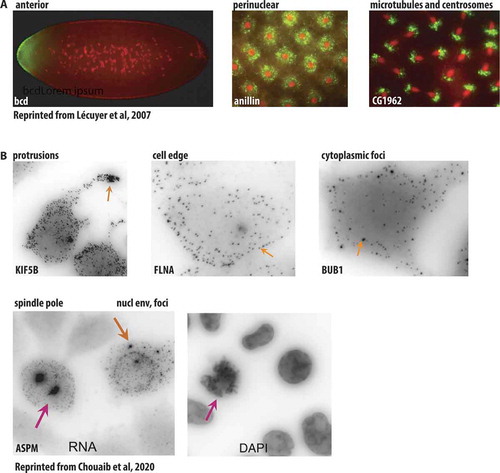
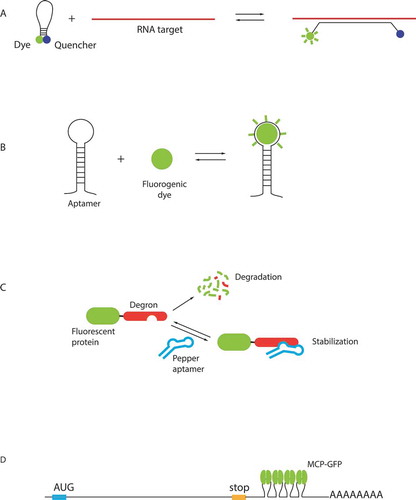
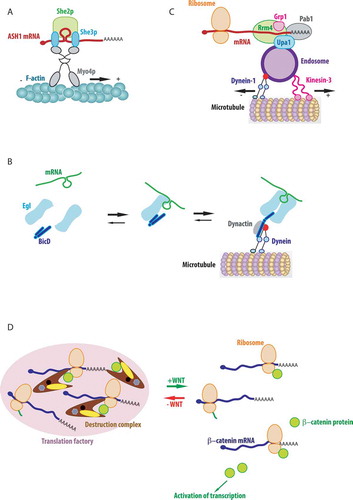
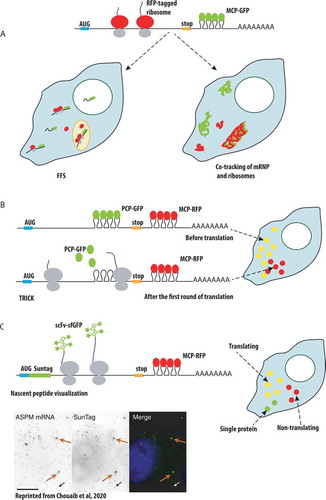
Figures & data
Femino AM, Fay FS, Fogarty K, et al. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. Tsanov N, Samacoits A, Chouaib R, et al. smiFISH and FISH-quant - a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016;44:e165. Chen KH, Boettiger AN, Moffitt JR, et al. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. Wu J, Zaccara S, Khuperkar D, et al. Live imaging of mRNA using RNA-stabilized fluorogenic proteins. Nat Methods. 2019;16:862–865. Bertrand E, Chartrand P, Schaefer M, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. Fusco D, Accornero N, Lavoie B, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol CB. 2003;13:161–167. Mofatteh M, Bullock SL. SnapShot: subcellular mRNA localization. Cell. 2017;169(178–178):e1. McClintock MA, Dix CI, Johnson CM, et al. RNA-directed activation of cytoplasmic dynein-1 in reconstituted transport RNPs. eLife. 2018;7:e36312. Higuchi Y, Ashwin P, Roger Y, et al. Early endosome motility spatially organizes polysome distribution. J Cell Biol. 2014;204:343–357. Chouaib R, Safieddine A, Pichon X, et al. A dual protein-mRNA localization screen reveals compartmentalized translation and widespread co-translational RNA targeting. Dev Cell. 2020;54(773–791):e5. Chin A, Lécuyer E. Translating messages in different neighborhoods. Dev Cell. 2020;54:691–693. Wu B, Buxbaum AR, Katz ZB, et al. Quantifying protein-mRNA interactions in single live cells. Cell. 2015;162:211–220. Katz ZB, English BP, Lionnet T, et al. Mapping translation “hot-spots” in live cells by tracking single molecules of mRNA and ribosomes. eLife. 2016;5:e10415. Halstead JM, Lionnet T, Wilbertz JH, et al. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 2015;347:1367–1671. Tanenbaum ME, Gilbert LA, Qi LS, et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646.