Figures & data
Figure 1. The suppressed expression of YB-1 entails YB-3 mRNA redistribution into polysomes without any effect on YB-3 stability. (A) Changes in the amount of YB-3 in HEK293T cells upon knockout or plasmid-induced expression of YB-1 detected by Western blotting; Rpl7 (ribosomal protein L7) is given as a control. (B) Changes in the amount of YB-1 in HEK293T cells upon YB-3 knockout detected by Western blotting; Rpl7 (ribosomal protein L7) is given as a control. (C) CHX-chase analyses of YB-3 degradation in HEK293T and HEK293TΔYB-1. The cells were incubated with CHX (100 µg/ml) and harvested at the indicated time points. Whole protein lysates were harvested, equilibrated to the total protein, and used in western blot analysis. p21/Waf was used as a positive control. Note that p21/Waf expression is very low in HEK293TΔYB-1 cells, as shown previously [Citation11]. (D) Distribution of YB-3 mRNA between polysomal and free mRNP fractions in HEK293T, HEK293T+YB-1, HEK293TΔYB-1, and HEK293T ΔYB-1+ YB-1 cells. The cells were scraped and lysed. Nuclei and mitochondria were removed by centrifugation, and cytosolic extracts were then spun through a 50% sucrose cushion at 100,000 rpm in a TLA-100 centrifuge (Beckman) for 13 min to separate postpolysomal supernatant from polysomes. Total RNA from postpolysomal supernatant and polysomal fractions (resuspended pellets) were extracted with TRIzol and subjected to qRT-PCR with YB-3 mRNA specific primers. Samples with added EDTA (for polysome dissociation into subparticles) served as a control for the presence of free mRNPs in polysomal fractions. The sum of relative mRNA YB-3 amount in free and polysomal mRNP fractions was taken to be 100%
![Figure 1. The suppressed expression of YB-1 entails YB-3 mRNA redistribution into polysomes without any effect on YB-3 stability. (A) Changes in the amount of YB-3 in HEK293T cells upon knockout or plasmid-induced expression of YB-1 detected by Western blotting; Rpl7 (ribosomal protein L7) is given as a control. (B) Changes in the amount of YB-1 in HEK293T cells upon YB-3 knockout detected by Western blotting; Rpl7 (ribosomal protein L7) is given as a control. (C) CHX-chase analyses of YB-3 degradation in HEK293T and HEK293TΔYB-1. The cells were incubated with CHX (100 µg/ml) and harvested at the indicated time points. Whole protein lysates were harvested, equilibrated to the total protein, and used in western blot analysis. p21/Waf was used as a positive control. Note that p21/Waf expression is very low in HEK293TΔYB-1 cells, as shown previously [Citation11]. (D) Distribution of YB-3 mRNA between polysomal and free mRNP fractions in HEK293T, HEK293T+YB-1, HEK293TΔYB-1, and HEK293T ΔYB-1+ YB-1 cells. The cells were scraped and lysed. Nuclei and mitochondria were removed by centrifugation, and cytosolic extracts were then spun through a 50% sucrose cushion at 100,000 rpm in a TLA-100 centrifuge (Beckman) for 13 min to separate postpolysomal supernatant from polysomes. Total RNA from postpolysomal supernatant and polysomal fractions (resuspended pellets) were extracted with TRIzol and subjected to qRT-PCR with YB-3 mRNA specific primers. Samples with added EDTA (for polysome dissociation into subparticles) served as a control for the presence of free mRNPs in polysomal fractions. The sum of relative mRNA YB-3 amount in free and polysomal mRNP fractions was taken to be 100%](/cms/asset/89715335-92f5-4728-bd54-a17cd758b320/krnb_a_1859243_f0001_b.gif)
Figure 2. The suppressed expression of YB-1 causes lower stability of YB-3 mRNA. Changes in stability (A-E) and synthesis (F) of some mRNAs in YB-1-null cells. (A-F) HEK293T and HEK293TΔYB-1 cells were incubated in DMEM with bromodeoxyuridine (BrU) for 24 h and then in standard medium for 0, 4, 8, or 12 h, followed by isolation of total RNA. This RNA was used for immunoprecipitation with antibodies against BrU. In the immunoprecipitate, selected mRNAs were detected by qRT-PCR. For each cell line, the mRNA amount at the 0 h point was taken to be 100%. Errors are 2 standard deviations. Two-tailed Student’s t-test was used to estimate the statistical significance. ***p < 0.001, *p < 0.05. (G) HEK293T and HEK293TΔYB-1 cells were incubated in DMEM with BrU for 24 h, followed by isolation of total RNA. This RNA was used for immunoprecipitation with antibodies against BrU. In the immunoprecipitate, selected mRNAs were detected by qRT-PCR. In HEK293T cells, the mRNA amount at the 0 h point was taken to be 100%. Errors are 2 standard deviations. Two-tailed Student’s t-test was used to estimate the statistical significance. ***p < 0.001, **p < 0.01
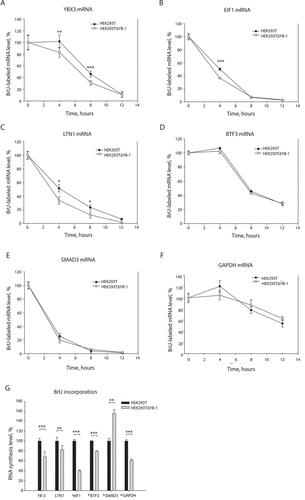
Figure 3. The 5ʹ and 3ʹ UTRs of YB-3 mRNA are required for regulation of translation and stability of reporter mRNAs in HEK293T cells. (A) Scheme of the used pNL2.2 plasmids. The HEK293T ΔYB-1 and HEK293TΔYB-1+ YB-1 cells were transfected using the pNL2.2 plasmid, encoding the reporter Nluc mRNA with a combination of UTRs from YB-3- and BTF3 mRNAs, and the pNL2.2BTF3_Fluc_BTF3 plasmid. 24 h after transfection, the cells were harvested and divided into two parts. One part was used to isolate total RNA for measuring the amounts of Nluc- and Fluc mRNAs by qRT-PCR. The other was used to determine Nluc and Fluc activities. (B) The NanoLuc activities (normalized to that of Fluc synthesized from pNL2.2BTF3_Fluc_BTF3) in HEK293T ΔYB-1 and HEK293TΔYB-1+ YB-1 cells for the pNL2.2 plasmids encoding reporter Nluc mRNAs with various UTRs of YB-3- and BTF3 mRNAs. Errors are 2 standard deviations. Two-tailed Student’s t-test was used to estimate the statistical significance; ns stands for non-significant. (C) The amounts of NanoLuc mRNAs (normalized to that of Fluc synthesized from pNL2.2BTF3_Fluc_BTF3) in HEK293ΔYB-1 T and HEK293TΔYB-1+ YB-1 cells for the pNL2.2 plasmids encoding reporter Nluc mRNAs with various UTRs of YB-3- and BTF3 mRNAs. Errors are 2 standard deviations. Two-tailed Student’s t-test was used to estimate the statistical significance. ***p < 0.001, *p < 0.05, ns stands for non-significant. (D) The activity of Nluc corresponding to the mRNA amount, derived from B and C. Errors are 2 standard deviations. Two-tailed Student’s t-test was used to estimate the statistical significance. **p < 0.01, *p < 0.05. (E) HEK293T ΔYB-1 and HEK293TΔYB-1+ YB-1 cells were transfected using the pNL2.2 encoding either reporter Nluc mRNA with UTR or YB-3 mRNA or BTF3 mRNA. 6 h after transfection, the reaction medium was supplemented with BrU for further 24 h incubation. Then, with the medium replaced by standard one, the cells were incubated for 0, 2, 4, or 8 h, followed by isolation of total RNA. This RNA was used for immunoprecipitation with antibodies against BrU. In the immunoprecipitate, YB-3_Nluc_YB-3 mRNA was detected by qRT-PCR. For each cell line, the mRNA amount at the 0 h point was taken to be 100%. Errors are 2 standard deviations. Two-tailed Student’s t-test was used to estimate the statistical significance. *p < 0.05
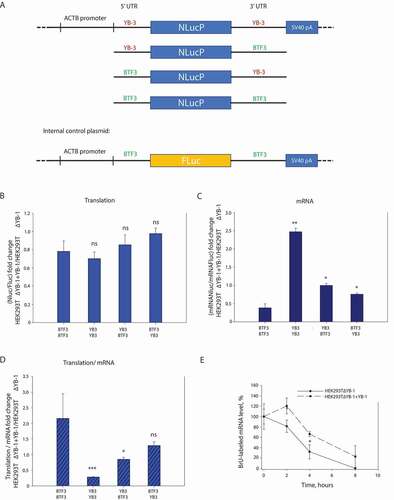
Figure 4. The 5ʹ UTR of YB-3 mRNA mediates regulation of translation of reporter mRNAs in a cell-free translation system. (A) Scheme of the used reporter mRNAs. 0.1 pmol C+A+ reporter
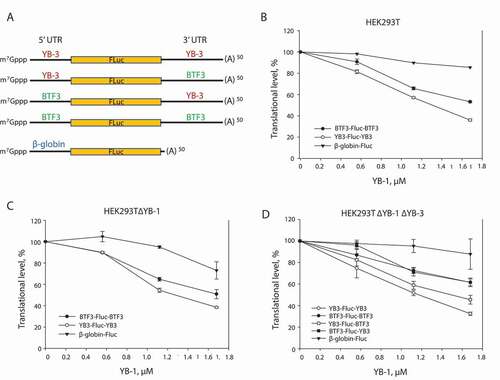
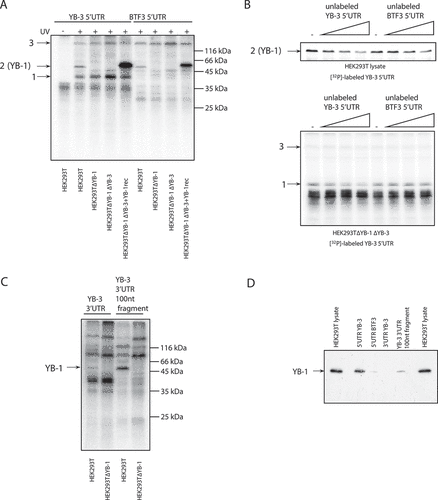
Figure 5. YB-1 binding to YB-3 mRNA UTRs. (A) Aliquots (5 μl) of HEK293T, HEK293T∆YB-1, HEK293T ∆YB-1 ∆YB-3 cell extracts or HEK293T ∆YB-1 ∆YB-3 cell extract supplemented with 1 µg of recombinant YB-1 were incubated for 15 min at 30 °C with 32P-labelled YB-3 mRNA 5′UTR (0.57 pmol) or BTF3 mRNA 5′UTR, UV cross-linked, treated with RNase A and MN, analysed by SDS-PAGE, and visualized by autoradiography. The numbers denote proteins specifically cross-linking withYB-3 mRNA 5′UTR. (B) Aliquots (5 μl) of HEK293T ∆YB-1 ∆YB-3 cell extract were incubated for 15 min at 30 °C with 32P-labelled YB-3 mRNA 5′UTR (0.57 pmol) in the presence of increasing amounts of unlabelled YB-3 mRNA 5′UTR or BTF3 mRNA 5′UTR (0.57, 1.14, and 2.28 pmol), UV cross-linked, treated with RNase A and MN, analysed by SDS-PAGE, and visualized by autoradiography. (С) Aliquots (5 μl) of HEK293TorHEK293T∆YB-1 were incubated for 15 min at 30 °C with 32P-labelled YB-3 mRNA 3′UTR (0.2 pmol) or YB-3 3ʹUTR 100 nt fragment (0.6 pmol), UV cross-linked, treated with RNase A and MN, analysed by SDS-PAGE, and visualized by autoradiography. (D) The proteins were adsorbed from HEK293T extract (50 µl) on Streptavidin Sepharose with 60 pmol of biotinylated YB-3 mRNA fragments or control BTF3 mRNA 5′UTR. The RNA-bound proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane. YB-1 was detected using appropriate polyclonal antibodies