Figures & data
Figure 1. LARP1 La-module confers poly(A) length protection and mRNA stabilization. A) Schematic representation of full-length LARP1 and two La-module constructs used in this study. Amino acid (aa) numbering is according to isoform 1 (1019 aa) for the full length and the shorter constructs. The PAM2 sequence is depicted in the linker ten amino acids following the end of the LaM (see in [Citation71]) and extending into the predicted RRM. RG indicates the position of the multiple Arg-Gly repeats [Citation46] (Tri-RG region [Citation45]); CR1 and CR2 indicate the conserved regions [Citation46]. B) Northern blot analysis of total RNA isolated from HEK293 cells transfected with the constructs indicated above the lanes. A single blot was probed for βG-ARE, GFP, and histone H2A mRNAs as indicated in panels i–iii). Bottom panel shows a vertically compacted image of the EtBr stained gel prior to transfer. C) Northern blot of RNAs from HEK293T LARP1-KO cells, as in B above. D) Quantitation of βG-ARE mRNA from duplicate northern blot data from HEK293T LARP1-KO cells. Transcript levels were normalized by VA1 small ncRNA. N = 2; error bars represent the spread. E) Western blot of total protein from the same cells used for RNA shown in panel E. F) The same blot as in C showing samples from a duplicate experiment, probed for the endogenous cellular rpL35 mRNA
![Figure 1. LARP1 La-module confers poly(A) length protection and mRNA stabilization. A) Schematic representation of full-length LARP1 and two La-module constructs used in this study. Amino acid (aa) numbering is according to isoform 1 (1019 aa) for the full length and the shorter constructs. The PAM2 sequence is depicted in the linker ten amino acids following the end of the LaM (see Fig. 5 in [Citation71]) and extending into the predicted RRM. RG indicates the position of the multiple Arg-Gly repeats [Citation46] (Tri-RG region [Citation45]); CR1 and CR2 indicate the conserved regions [Citation46]. B) Northern blot analysis of total RNA isolated from HEK293 cells transfected with the constructs indicated above the lanes. A single blot was probed for βG-ARE, GFP, and histone H2A mRNAs as indicated in panels i–iii). Bottom panel shows a vertically compacted image of the EtBr stained gel prior to transfer. C) Northern blot of RNAs from HEK293T LARP1-KO cells, as in B above. D) Quantitation of βG-ARE mRNA from duplicate northern blot data from HEK293T LARP1-KO cells. Transcript levels were normalized by VA1 small ncRNA. N = 2; error bars represent the spread. E) Western blot of total protein from the same cells used for RNA shown in panel E. F) The same blot as in C showing samples from a duplicate experiment, probed for the endogenous cellular rpL35 mRNA](/cms/asset/bb110ceb-915e-4f00-8534-06292115723a/krnb_a_1860376_f0001_b.gif)
Figure 2. PAM2-dependent stable binding to PABP by the isolated La-modules. A) Western blot showing extracts used for immunoprecipitation (IP) by anti-FLAG IgG (input, lanes 1–7) and products of IP in the absence (lanes 8–14) and presence of RNase I (lanes 15–21). The extracts were isolated from HEK293 cells transfected with the Flag-tagged constructs indicated above the lanes. The antibodies used to detect proteins on the blots are indicated to the right of the panels. The bottom panel shows an EtBr stained gel of a fraction of the extract after the IP and processed for RNA purification. B) Quantitation of the results of duplicate experiments, the levels of PABP that co-IP with La-module 205–509 were set to 100% in both mock and +RNase I treated extract. The error bars represent the spread
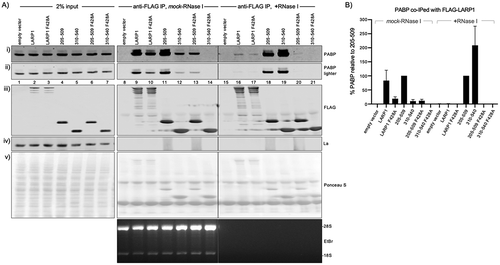
Figure 3. The F428A PAM2 mutation does not inhibit oligo(A)-binding by the La-module. A) SDS PAGE analysis of the purified recombinant La-module 310–540 and La-module 310–540 F428A proteins used for RNA binding. The expected mass is 29.9 kDa; molecular weight markers (MWM) are indicated. B) RNA binding by Electrophoretic mobility shift analysis (EMSA). The positions of the free 32P-r(A)25 RNA and the RNA-protein (RNP) complex are indicated. The protein concentrations ranged from 0.03 μM to 7.5 μM indicated above the lanes as triangle ramps
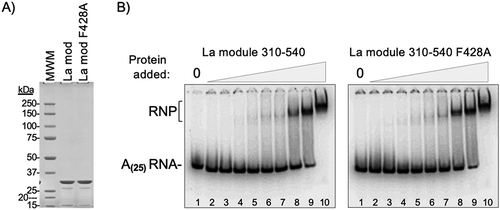
Figure 4. Characterization of a LARP1 PAM2 sequence that binds MLLE with high affinity. A) Multiple sequence alignment reveals distinctive features of the LARP1 PAM2 sequence. ClustalW alignment performed using MacVector (Methods). The PAM2 consensus numbering is indicated above the alignment. A separate alignment of the human TNRC6A/B/C sequences, offset according to Kozlov et al [Citation55] (see text) is below the main alignment; it includes the high-affinity MLLE-binding peptide of GW182/TNRC6C that was characterized in detail [Citation55]. B) Sequence LOGOs derived from the mammalian, opossum, and frog PAM2 LARP1 and LARP1B sequences compiled by Deragon [Citation46]. C) 15N-1H NMR correlation spectra (left) and ITC thermograms (right) of the wild-type PAM2 peptide (TDFSQLLNCPEFVPRQHYQKE) titrated into the PABP MLLE domain. Arrows in the NMR spectra show the shifts of two glycine signals upon PAM2 binding. The pattern of chemical shift changes in the MLLE domain is typical for high-affinity PAM2 binding. D) NMR and ITC titrations with the F428A mutant PAM2 peptide (TDFSQLLNCPEAVPRQHYQKE) show no binding. E) Titrations with a recombinant LARP1 fragment 399–540 containing both the PAM2 motif and full predicted RRM domain show slightly reduced binding affinity relative to the isolated PAM2 peptide. Nonetheless, the patterns of spectral changes are identical (arrows), which confirms that both the PAM2 peptide and PAM2-RRM domain bind to the same site on the MLLE domain
![Figure 4. Characterization of a LARP1 PAM2 sequence that binds MLLE with high affinity. A) Multiple sequence alignment reveals distinctive features of the LARP1 PAM2 sequence. ClustalW alignment performed using MacVector (Methods). The PAM2 consensus numbering is indicated above the alignment. A separate alignment of the human TNRC6A/B/C sequences, offset according to Kozlov et al [Citation55] (see text) is below the main alignment; it includes the high-affinity MLLE-binding peptide of GW182/TNRC6C that was characterized in detail [Citation55]. B) Sequence LOGOs derived from the mammalian, opossum, and frog PAM2 LARP1 and LARP1B sequences compiled by Deragon [Citation46]. C) 15N-1H NMR correlation spectra (left) and ITC thermograms (right) of the wild-type PAM2 peptide (TDFSQLLNCPEFVPRQHYQKE) titrated into the PABP MLLE domain. Arrows in the NMR spectra show the shifts of two glycine signals upon PAM2 binding. The pattern of chemical shift changes in the MLLE domain is typical for high-affinity PAM2 binding. D) NMR and ITC titrations with the F428A mutant PAM2 peptide (TDFSQLLNCPEAVPRQHYQKE) show no binding. E) Titrations with a recombinant LARP1 fragment 399–540 containing both the PAM2 motif and full predicted RRM domain show slightly reduced binding affinity relative to the isolated PAM2 peptide. Nonetheless, the patterns of spectral changes are identical (arrows), which confirms that both the PAM2 peptide and PAM2-RRM domain bind to the same site on the MLLE domain](/cms/asset/c56d6206-d762-4b8d-b3be-541bce134082/krnb_a_1860376_f0004_c.jpg)
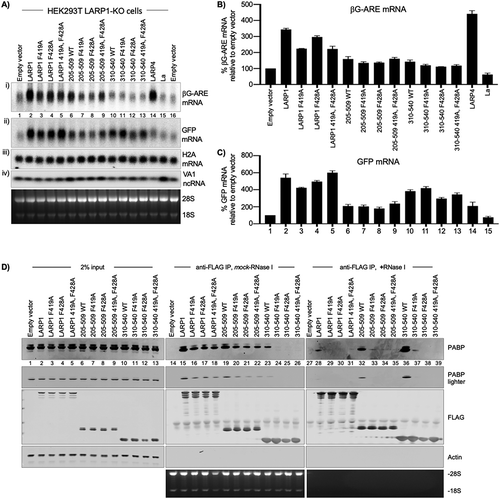
Figure 5. LARP1 PAM2 position-1 F419 is critical for functional and physical PABP interaction. A) Northern blot analysis of total RNA isolated from HEK293T LARP1-KO cells transfected with the constructs above the lanes. A single blot was probed for βG-ARE, GFP, and histone H2A mRNAs as well as the VA1 small ncRNA as indicated for panels i–iv). The bottom panel shows an image of the EtBr stained gel prior to transfer. B & C) Quantification of duplicate northern blot data for βG-ARE mRNA and GFP mRNA from the HEK293T LARP1-KO cells. Transcript levels were normalized by VA1 small ncRNA. N = 2; error bars represent the spread. D) Western blot of protein isolated from HEK293 cells used for immunoprecipitation (IP) of transfected Flag-tagged LARP1 constructs indicated above the lanes and developed using antibodies to proteins or epitopes indicated to the right of the panels. The bottom panel shows an image of the EtBr stained gel (see text)