Figures & data
Table 1. List of RiboMethSeq detected 2ʹ-O-methylation sites in A. thaliana 5.8S, 18S and 25S rRNAs. Mapped rRNA Nm sites (‘Mapped site’) are compared with previously known locations (‘Position 3D rRNAdb’ and associated C/D snoRNA [Citation22]). Conservation of rRNA modification sites with human (‘Position in human’) and yeast (‘Position in yeast’) profile was determined based on 2D rRNA structure and sequence [Citation8]. Human rRNA sites located in the same structural context, but not strictly conserved, are shown in grey. Newly detected snoRNA guides corresponding to the modified positions are shown (‘Assigned snoRNA’). The absence of identified snoRNA guide is shown in grey. Reduction of 2ʹ-O-methylation in nuc1-2 plants is shown in columns ‘ScoreC nuc1’ with corresponding p-value. Asterisks ****(≤0.0001), ***(≤0.001), **(≤ 0.01), *(≤0.05) represent significance level and ns, non-significant value ≥ 0.05
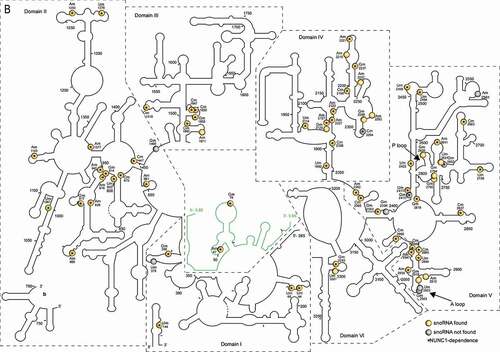
Figure 1. Predicted 18S, 5.8S and 25S rRNA structures with mapped 2ʹ-O-methylations sites (A) 18S and (B) 5.8S/25S rRNA structures were generated on the basis of 2D structural predictions [Citation24]. 2ʹ-O-methylated sites in yellow circles or grey circles show rRNA sites with or without matching C/D snoRNAs (respectively). Star inside the circles indicates observed dependency of rRNA 2ʹ-O-methylation on NUC1 gene expression. In B, the 5.8S rRNA structure is coloured in green and two 25S fragments (5ʹ-fragment and 3ʹ-fragment, a and b, respectively) are split in the main structure for clarity. The 18S (5ʹ, central, 3ʹMajor and 3ʹMinor) and 25S (I–VI) rRNA domains are designated according to [Citation1]. P and A loop are indicated by arrows
![Figure 1. Predicted 18S, 5.8S and 25S rRNA structures with mapped 2ʹ-O-methylations sites (A) 18S and (B) 5.8S/25S rRNA structures were generated on the basis of 2D structural predictions [Citation24]. 2ʹ-O-methylated sites in yellow circles or grey circles show rRNA sites with or without matching C/D snoRNAs (respectively). Star inside the circles indicates observed dependency of rRNA 2ʹ-O-methylation on NUC1 gene expression. In B, the 5.8S rRNA structure is coloured in green and two 25S fragments (5ʹ-fragment and 3ʹ-fragment, a and b, respectively) are split in the main structure for clarity. The 18S (5ʹ, central, 3ʹMajor and 3ʹMinor) and 25S (I–VI) rRNA domains are designated according to [Citation1]. P and A loop are indicated by arrows](/cms/asset/adb173af-9895-4f41-8746-56646fef13d5/krnb_a_1869892_f0001a_oc.jpg)
Figure 2. Characterization of FIB2-YFP snoRNP (A) Arabidopsis C/D-box protein orthologues (left) and C/D snoRNA (right) detected in the FIB2-YFP IP fractions using LC-MS/MS and RNA-seq approaches. The number of peptides and % of protein coverage for each protein is shown and the number of predicted 25S, 18S and 5.8S rRNA targets identified in the 5ʹ and 3ʹ antisense sequences of 141 C/D snoRNA. (B) Representation of 6 novel C/D snoRNA detected in the FIB2-IP fraction. C and D boxes are boxed. These C/D snoRNA do not contain a matching rRNA target sequence
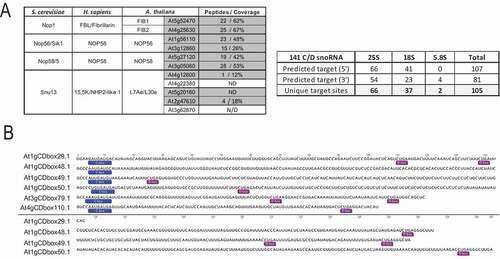
Table 2. List of C/D box snoRNAs identified from RNA-seq data. The number of C/D snoRNA reads are provided for each Co_IP fraction. Are indicated, rRNA positions mapped as 2ʹ-O-methylated in this study, when predicted to be guided by a snoRNA and the names of known C/D snoRNA in databases Are highlighted in red snoRNAs with predicted rRNA methylation sites but not detected by RiboMethSeq. ID with an asterisk identifies a snoRNA which is dicistronic with a tRNA. ID with two asterisks represents a snoRNA which is polycistronic with a miRNA
Figure 3. Mfold structure of At3gCDbox1011 and At5gDbox123.1 identified in silico and alignment with 25S rRNA sequences. Arrows show mapped rRNA sites Gm2794 and Cm1847 in the 25S and their counterpart in the snoRNA. Blue and red bars underline potential C and D boxes sequences
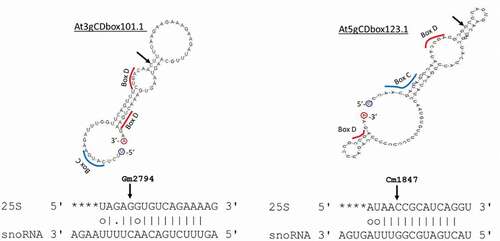
Figure 4. Heat map representation of rRNA 2ʹ-O-methylation in Arabidopsis. Differential methscore levels for 115 rRNA 2ʹ-O-methylated sites observed in three Col-0 and three nuc1-2 biological replicates (R1-R3) are shown. The rRNA sites are clustered according to hclust/ward.D2 method. Arrow heads show rRNA sites without associated guiding snoRNA. The colour key, histogram and values are shown
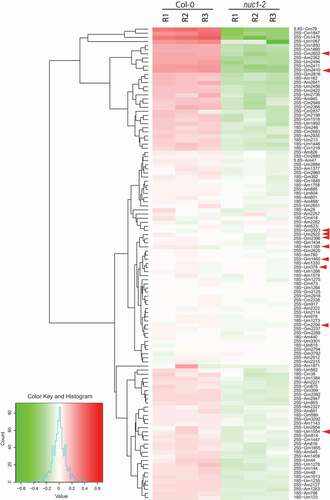
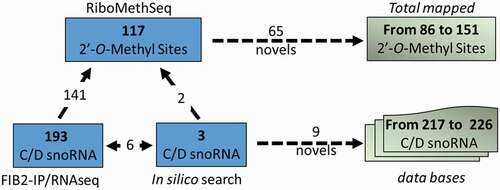
Figure 5. Illustration shows the number of 2ʹ-O-methylated sites mapped by RiboMethSeq (117) and C/D snoRNA detected in the FIB2 immunoprecipitated fraction (193) and/or identified by bioinformatics search (9). From these analyses we account 65 first time mapped 2ʹ-O-methylated sites and 9 novel C/D snoRNAs, increasing the number of 2ʹ-O-methylated mapped sites and C/D snoRNA identified and listed in Arabidopsis databases to 151 and 226 respectively