Figures & data
Figure 1. Schematic representation of components of U1 small nuclear ribonucleoprotein (snRNP) complex. The U1 small nuclear RNA (snRNA) is illustrated in black with secondary structures. m3Gppp shows U1 snRNA trimethyl cap. U1-specific proteins, including U1-70 K (green), U1-A(red) and U1-C(yellow), are shown with ellipses. Their positions correspond to their specific inter-molecular interactions (U1-70 K interacts with SL1; U1-A interacts with SLII; U1-C does not interact with U1 snRNA, while it is close to U1 snRNA 5ʹ-end and has contact with U1-70 K and Sm protein) . Sm proteins are shown with circles, they form a core and bind AUUUGUG sequence within U1 snRNA
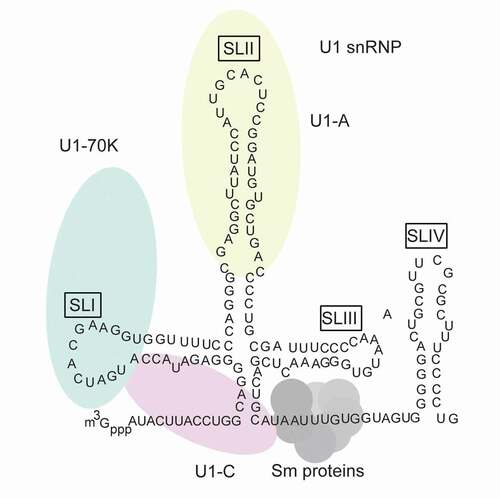
Figure 2. U1 snRNP inhibits cryptic PAS processing. U1 snRNP associates with pre-mRNA at many 5ʹ-SS or 5ʹ-SS like sites through its free U1 snRNA 5ʹ-end, which may inhibit the 3ʹ-end processing of downstream PAS and prevent PCPA. In addition to canonical PASs at 3ʹ-UTR, pre-mRNAs harbour many intronic cryptic PASs. Based on current knowledge, mRNA 3ʹ-end processing is carried out within mRNA 3ʹ-end processing complex, which includes trans-acting 3ʹ processing factors and core PAS cis-elements. It is generally believed that 3ʹ-UTR canonical PASs and intronic cryptic PASs share similar core cis-elements (AAUAAA motif, U/GU-rich sequences etc.), and the specific protein–RNA interactions within the 3ʹ processing complex are crucial for this process. (CPSF: cleavage and polyadenylation specificity factor; CstF: cleavage stimulation factor; CFIm: cleavage factor Im; CFIIm: cleavage factor IIm; PAP: poly(A) polymerase)
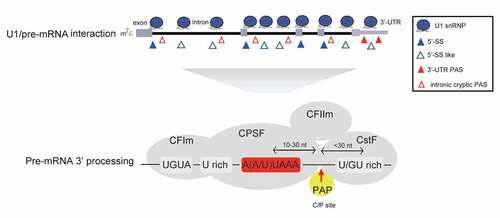
Figure 3. U1 snRNP or U1 component inhibits pre-mRNA 3ʹ-end processing in the context of multiple PASs. (A) U1 snRNP inhibits the cleavage step of mRNA 3ʹ-end processing for the PASs of indicated genes. 5ʹ-SS and C/P site are indicated with filled rectangle and arrow, respectively, and their orientations and distances are also indicated. (B) U1 snRNP inhibits the polyadenylation step in the context of BPV/L3 PASs, which is mediated by direct interaction between U1-70 K and poly(A) polymerase (PAP). (C) U1A inhibits the cleavage or polyadenylation step of mRNA 3ʹ-end processing in the context of indicated PASs. Cleavage/polyadenylation (C/P) sites are indicated with red arrows
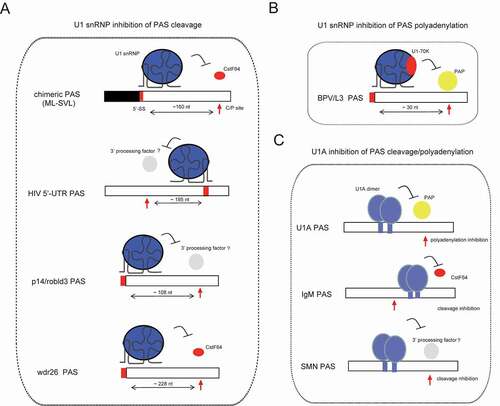
Figure 4. Schematic representation of reported models in elucidating U1 telescripting mechanism. (A) U1–CPAF (U1–cleavage and polyadenylation factors) model. Under physical conditions, U1–CPAF forms a complex near intronic cryptic PASs region, and its PAS processing activity is inhibited. In contrast, U1–CPAF becomes active upon U1 AMO treatment, resulting in efficient C/P at intronic PASs. This U1 AMO-mediated activation of U1–CPAF may be at least partially caused by the altered CPAF components. For instance, CFIm68, instead of CFIm59, accumulates in CPAF upon U1 AMO treatment. (B) FUS–U1 co-inhibition model. FUS and U1 snRNP form a complex, bind sequences upstream of intronic cryptic PASs and consequently inhibit the downstream PAS processing. This inhibitory effect may be caused by impaired recruitment of core 3ʹ processing factors, such as CPSF160
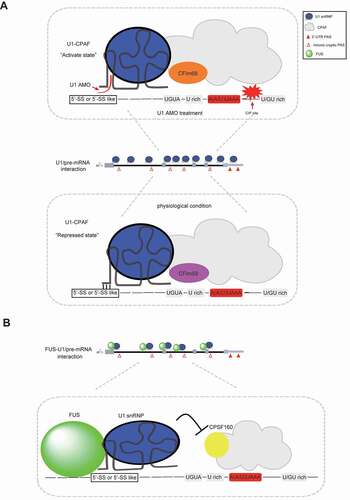
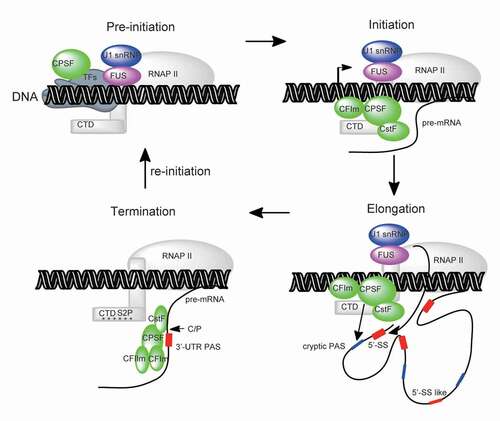
Figure 5. Recruitment of core mRNA processing factors (mRNA 3ʹ-end processing factors, U1 snRNP) in the context of co-transcriptional mRNA processing. CPSF connects with transcription pre-initiation through general transcription factors (TFIID) and transfers to RNA polymerase II (RNAP II) during transcription initiation. Other mRNA processing factors (CstF, CFIm and U1 snRNP/FUS) may also direct bind RNAP II during transcription pre-initiation/initiation/elongation. These stepwise co-transcriptional events may increase local concentration of mRNA processing factors and thereby promote the mRNA processing efficiency once RNA is transcribed. mRNA termination is coupled with mRNA 3ʹ-end processing at canoical PAS within 3ʹ-UTR, during which fully assembled 3ʹ processing complex recognizes and processes PAS efficiently