Figures & data
Figure 1. Workflow diagrams of PBL and CBRPP
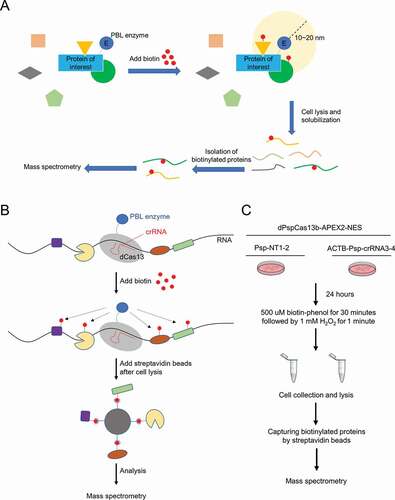
Table 1. crRNAs used in this paper
Table 2. qPCR primers used in this paper
Figure 2. dRfxCas13d is not suitable for CBRPP to study RNA-protein interactions
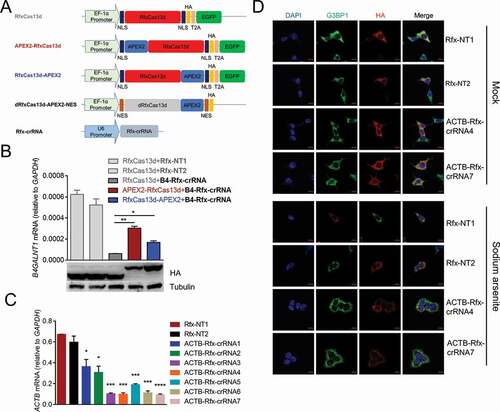
Figure 3. Transient transfection of dPspCas13b-APEX2-NES to identify the RBPs of ACTB mRNA
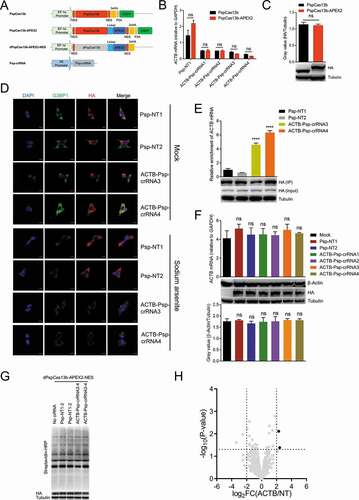
Figure 4. Identifying the RBPs of ACTB mRNA in cells inducibly expressing dPspCas13b-BioID2/TurboID/APEX2-NES
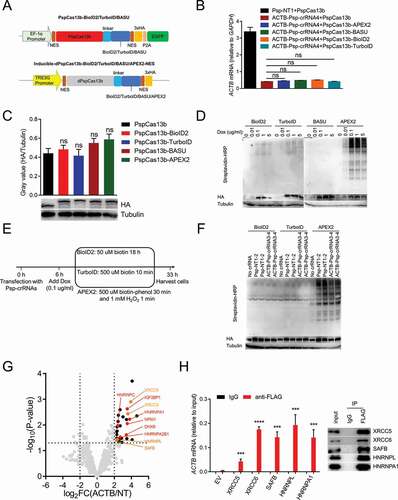
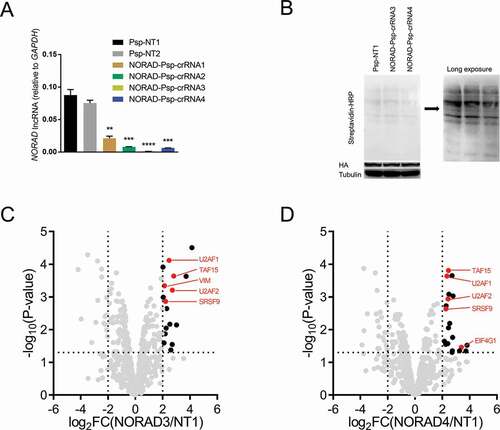
Figure 5. Identifying the RBPs of NORAD using CBRPP