Figures & data
Figure 1. Sequence, structure and activity of the P. polymyxa pistol ribozyme. (A) Sequence and secondary structure of the P. polymyxa pistol ribozyme. Nucleotides in red correspond to the highly conserved positions from the consensus model [Citation14]. Loop 3 (nucleotides coloured grey) is absent in the bimolecular construct (see Supplementary Fig. 1A for details). Altered nucleotides in the mutant construct are boxed. P1-, P2- and P3-stems are indicated. Cleavage site is indicated by a black arrowhead. (B) Self-cleavage activity of the P. polymyxa pistol ribozyme. Internally labelled cleavage products were separated by denaturing 20% PAGE. The full-length ribozyme (FL) and cleavage products (5ʹ clv; 3ʹ clv) are indicated by black arrowheads. T1: RNase T1-ladder; OH−: partial alkaline digest-ladder (see Materials and methods for details). WT: wild-type ribozyme; MT: mutant ribozyme. (C) Cleavage activity of the P. polymyxa bimolecular pistol ribozyme construct. The 32P-labelled substrate RNA (5 nM) was incubated for the times indicated in the presence (+) or absence (-) of 20 mM MgCl2 and unlabelled enzyme RNA (100 nM) (see Materials and methods for details). Cleavage products were separated by denaturing 20% PAGE. The full-length substrate (sub) and 5ʹ-cleavage product (5ʹ clv) are denoted
![Figure 1. Sequence, structure and activity of the P. polymyxa pistol ribozyme. (A) Sequence and secondary structure of the P. polymyxa pistol ribozyme. Nucleotides in red correspond to the highly conserved positions from the consensus model [Citation14]. Loop 3 (nucleotides coloured grey) is absent in the bimolecular construct (see Supplementary Fig. 1A for details). Altered nucleotides in the mutant construct are boxed. P1-, P2- and P3-stems are indicated. Cleavage site is indicated by a black arrowhead. (B) Self-cleavage activity of the P. polymyxa pistol ribozyme. Internally labelled cleavage products were separated by denaturing 20% PAGE. The full-length ribozyme (FL) and cleavage products (5ʹ clv; 3ʹ clv) are indicated by black arrowheads. T1: RNase T1-ladder; OH−: partial alkaline digest-ladder (see Materials and methods for details). WT: wild-type ribozyme; MT: mutant ribozyme. (C) Cleavage activity of the P. polymyxa bimolecular pistol ribozyme construct. The 32P-labelled substrate RNA (5 nM) was incubated for the times indicated in the presence (+) or absence (-) of 20 mM MgCl2 and unlabelled enzyme RNA (100 nM) (see Materials and methods for details). Cleavage products were separated by denaturing 20% PAGE. The full-length substrate (sub) and 5ʹ-cleavage product (5ʹ clv) are denoted](/cms/asset/b632b5fb-f20b-45d1-9288-32a94a33be37/krnb_a_1874706_f0001_oc.jpg)
Figure 2. Metal ion dependency of the P. polymyxa pistol ribozyme cleavage. (A) Pistol ribozyme cleavage assays with the P. polymyxa bimolecular construct incubated for 30 min in the absence (-) or presence of various 1 mM divalent metal ions. (B) Dependence of the P. polymyxa pistol ribozyme cleavage rate on the pKa of the hydrated divalent metal ion. Six divalent metal ions at the concentration of 1 mM, pH 6.0 and 2 M NaCl [reaction conditions according to Citation18] were tested. pKa values are taken from Citation30. (C) Reactions of the P. polymyxa bimolecular construct in the absence (-) or presence (+) of 5 mM cobalt hexammine chloride [Co(NH3)6Cl3] or MgCl2 for 60 min in the presence of 5 mM EDTA. (D) Pistol ribozyme cleavage assays with the P. polymyxa bimolecular construct incubated for 60 min in the absence (-) or presence of various 1 M monovalent metal ions. To chelate contaminating divalent metal ions, 30 mM EDTA was added to these reactions. For the positive control (PC), 25 mM MgCl2 was added
![Figure 2. Metal ion dependency of the P. polymyxa pistol ribozyme cleavage. (A) Pistol ribozyme cleavage assays with the P. polymyxa bimolecular construct incubated for 30 min in the absence (-) or presence of various 1 mM divalent metal ions. (B) Dependence of the P. polymyxa pistol ribozyme cleavage rate on the pKa of the hydrated divalent metal ion. Six divalent metal ions at the concentration of 1 mM, pH 6.0 and 2 M NaCl [reaction conditions according to Citation18] were tested. pKa values are taken from Citation30. (C) Reactions of the P. polymyxa bimolecular construct in the absence (-) or presence (+) of 5 mM cobalt hexammine chloride [Co(NH3)6Cl3] or MgCl2 for 60 min in the presence of 5 mM EDTA. (D) Pistol ribozyme cleavage assays with the P. polymyxa bimolecular construct incubated for 60 min in the absence (-) or presence of various 1 M monovalent metal ions. To chelate contaminating divalent metal ions, 30 mM EDTA was added to these reactions. For the positive control (PC), 25 mM MgCl2 was added](/cms/asset/1963f7d9-6403-4b4d-9c75-b00e4e0d2230/krnb_a_1874706_f0002_b.gif)
Figure 3. Kinetic characteristics of the P. polymyxa pistol ribozyme cleavage. (A) Effect of Mg2+ on the rate of P. polymyxa pistol ribozyme cleavage. The log-log plot of kobs values at pH 6.0 versus MgCl2 concentrations ranging from 0.1 mM to 25 mM is shown. (B) Effect of pH on the rate of P. polymyxa pistol ribozyme cleavage. The log-log plot of kobs values at the MgCl2 concentration of 1 mM versus pH values ranging from 5.25 to 8.75 is depicted
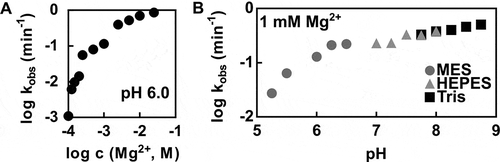
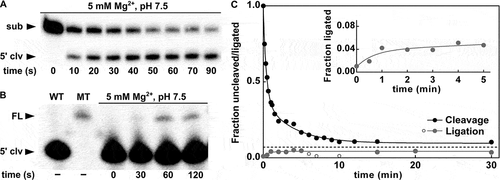
Figure 4. Ligation activity of the P. polymyxa pistol ribozyme. (A) Kinetic assay using the P. polymyxa bimolecular ribozyme construct for cleavage (see Materials and methods for details) at the MgCl2 concentration of 5 mM and 30 mM Tris (pH 7.5). The corresponding time points are indicated. The cleavage products were separated by denaturing 20% PAGE. The full-length substrate (sub) and 5ʹ-cleavage product (5ʹ clv) are denoted. (B) Kinetic ligation assay of the P. polymyxa pistol ribozyme at the MgCl2 concentration of 5 mM and 30 mM HEPES (pH 7.5). 15% PEG8000 was used as an additive to improve the assay quantification. The corresponding time points are indicated. The ligation products were separated by denaturing 10% PAGE. The full-length ribozyme (FL) and 5ʹ-cleavage product (5ʹ clv) are denoted. As a positive control, hot transcription of the wild-type (WT) and mutant (MT) P. polymyxa pistol ribozyme was performed. (C) Comparison of the cleavage and ligation activities of the P. polymyxa pistol ribozyme. Approach to equilibrium at 5 mM Mg2+ and pH 7.5 is shown. The dashed line represents the mean value of the fraction of the full-length ribozyme (ligation) or uncleaved substrate (cleavage) at equilibrium, feq = 0.07. Open grey circles indicate cleavage of the ligated ribozyme, which was detected during the ligation assay. The inset shows a zoom-in of the region between 0 min and 5 min of the ligation reaction